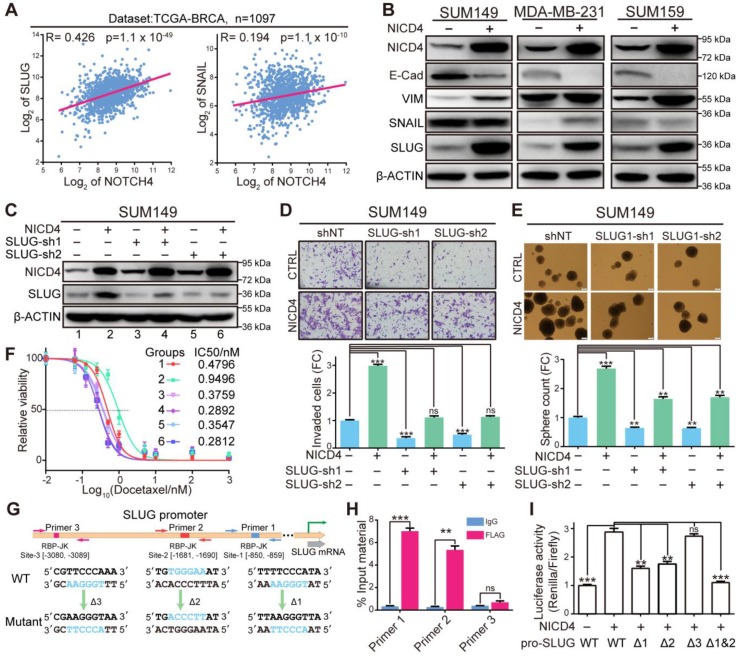
Figure 5.
NICD4 directly upregulated SLUG expression to induce EMT. (A) Gene expression correlation between NOTCH4 and SLUG or SNAIL was plotted via R2 (https://hgserver1.amc.nl/cgi-bin/r2/main.cgi) using dataset from TCGA-BRCA. (B) Markers for EMT (VIM, SNAIL and SLUG) and MET (E-Cad) were detected at protein level in NICD4-overexpressing SUM149, MDA-MB-231, and SUM159. (C) SLUG was interfered by shRNA in NICD4 overexpressing cells or control cells. The knockdown efficiency was determined by western blotting, using β-ACTIN as loading control. (D-F) We then determined (D) invasion ability by transwell assay, (E) mammosphere formation capability and (F) chemosensitivity via MTT assay after SLUG knockdown in SUM149 cells, the groups represented by numbers in (F) were the same as in (C). (G) Schematic depiction of the SLUG promoter with putative binding sites (colored rectangles) indicated. Primer sets for ChIP analyses were indicated by the arrows and the designed mutants were shown below. (H) ChIP and qRT-PCR were subsequently performed. The results of each primer were expressed as % Input material. IgG was used as control. (I) Dual-luciferase reporter assays of the SLUG promoter region. The relative luciferase signal ratio (Relina Luc. /Firefly Luc.) was further normalized to that of the group transfected with control vector. Triple independent experiments were carried out and representative results were shown. ns=not significant, ** P<0.01, *** P<0.001.