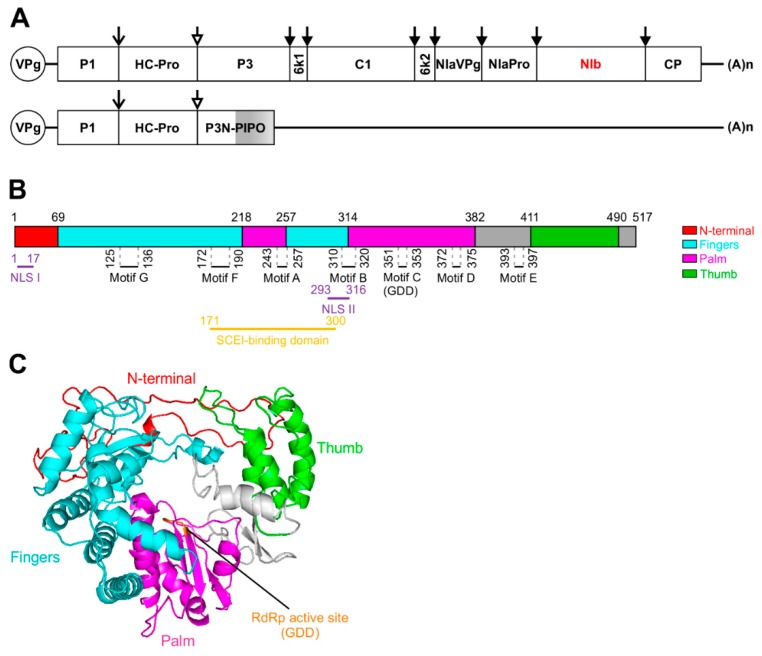
Figure 1.
Schematic representation of the Turnip mosaic virus (TuMV) genome and the predicted structure of the TuMV nuclear inclusion b (NIb). (A) Schematic representation of the TuMV genome. The circle represents the genome-linked viral protein VPg, and (A)n represents the poly(A) tail. The open reading frame (ORF) is indicated as a long box. Mature proteins resulting from polyprotein processing are indicated by smaller boxes. PIPO derived from a frameshift on the P3 cistron is indicated as a grey box. P1 and HC-Pro release themselves by auto-proteolytic cleavage at their own C-termini. Other cleavages are processed by NIaPro. The mature proteins include coat protein (CP), nuclear inclusion b (NIb), which is the viral RNA-dependent RNA polymerase (RdRp), and two viral suppressors of RNA silencing (VSRs), HC-Pro and VPg. For clarity, the relative sizes of the mature proteins are not drawn to scale. (B) Schematic representation of TuMV NIb showing the predicted locations of RdRp signature motifs A to G. The locations of motifs A to G was determined by protein sequence alignment of TuMV NIb and RHDV RdRp. NLSs, the GDD motif, and the SCE1 binding domain are indicated. (C) Ribbon diagram of TuMV NIb predicted by the I-TASSER server. The TuMV NIb protein (NP_734221.1) sequence was submitted to the I-TASSER server for 3D structure prediction and the Rabbit hemorrhagic disease virus (RHDV) RdRp (PDB ID:1KHV) was identified as the most homologous protein in the PDB library. The typical RdRp fingers, palm, and thumb domains are colored cyan, magenta, and green, respectively, and the N-terminal domain is colored red. The RdRp active site is colored orange.