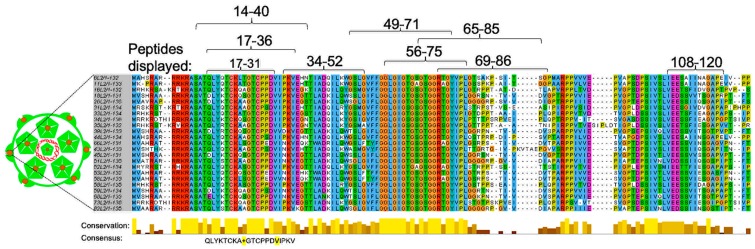
Figure 3.
Sequence alignment of L2 (aa 1–135) from different HPV types. Sequence alignment was done using sequences from both high-risk (HPVs 16, 18, 26, 31, 33, 34, 35, 39, 45, 51–53, 56, 58, 59, 66, 68, 73, and 82) and low-risk HPV types (HPVs 6, 11, 43, and 44). Alignment was done using Jalview software. Residues that are highly conserved among different HPV types are highlighted, below the alignment, in gold bars. A consensus sequence of amino acid 17–36 from different HPV types is shown below the bars. Amino acid residues (in the consensus sequence) that differ from HPV16 L2 are highlighted in yellow background. The numbers above sequence alignment represent peptides (amino acid residues) that have been displayed on different VLP platforms (see Table 1 and Table 2 for details).