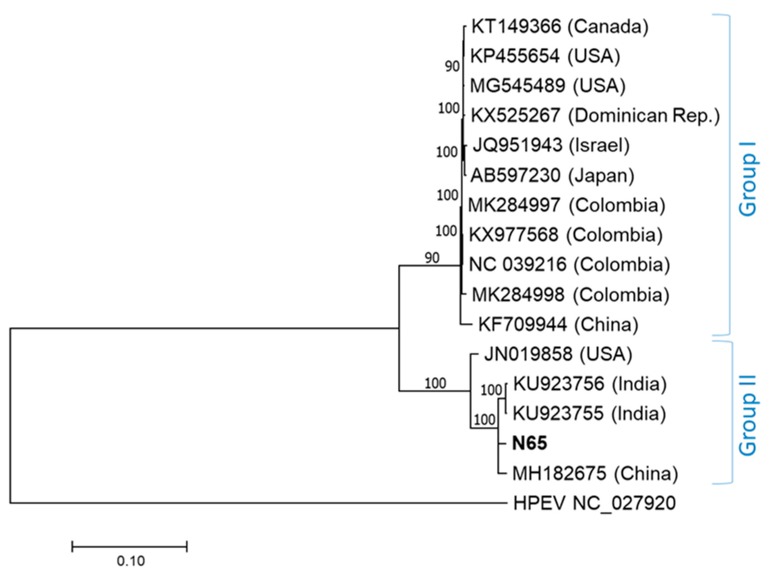
Figure 1.
Phylogenetic tree generated on complete nucleotide genome sequences of bell pepper endornavirus (BPEV) isolates. The phylogenetic analysis was inferred using maximum likelihood (ML) based on the General Time Reversible (GTR) model selected as the best-fit model of nucleotide substitution based on Bayesian information criterion (BIC) as implemented in MEGA 7. Isolates are identified by their GenBank accession numbers and country of origin. The Slovak isolate N65 sequenced in the present study is highlighted in bold. Hot pepper endornavirus (HPEV) was used as an outgroup. The scale bar indicates a genetic distance of 0.1. Bootstrap values higher than 70% (1000 bootstrap resamplings) are indicated.