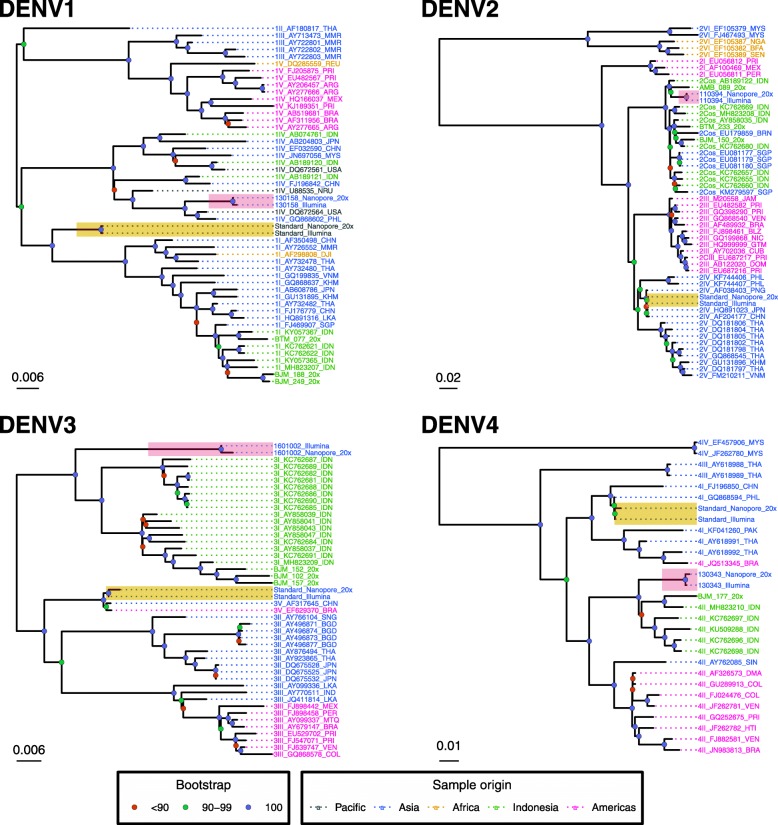
Fig. 4.
Phylogenetic analysis of Illumina- and Nanopore-generated consensus sequences. Bootstrap phylogenies of complete DENV coding regions were constructed using Nanopore and Illumina consensus sequences and a selection of genotype reference sequences. Nanopore consensus sequences were generated for all samples using the short amplicon approach, with regions below 20X coverage depth masked. Illumina consensus sequences were generated for the RNA standards and Pilipino samples using the long-amplicon approach. Sequence names are coloured to denote geographical origin, and internal nodes of the tree are coloured to demonstrate bootstrap values (blue = 100%, green = 90–99% and red = < 90%). Monophyletic clades formed by the Nanopore and Illumina-generated consensus sequences are highlighted in yellow for the RNA standard samples, and red for the Pilipino clinical samples