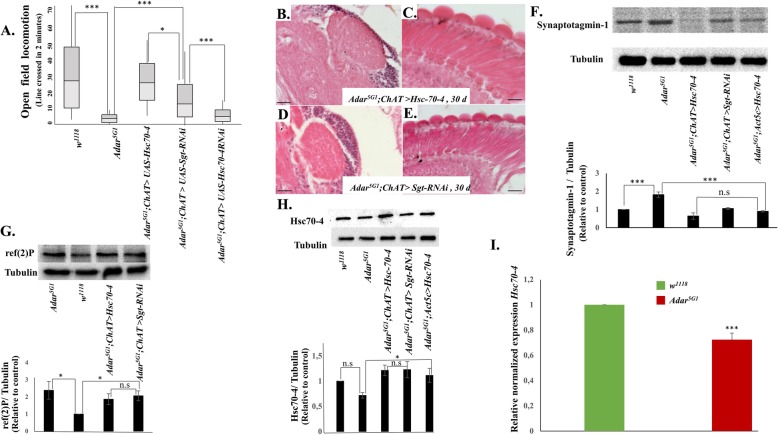
Fig. 6.
Increased Hsc70-4 suppresses Adar5G1 mutant phenotypes. a Rescue of Adar5G1 mutant open field locomotion defects in Adar5G1; ChAT > Hsc70-4 and Adar5G1; ChAT > Sgt RNAi flies with increased endosomal microautophagy. n ≥ 10. b Representative images of MB calyx (× 40) and c retina in 30-day Adar5G1; ChAT>Hsc70-4 (× 40).d Representative images of MB calyx (× 40) and e retina in 30-day Adar5G1; ChAT>SgtRNAi (× 40). f Immunoblot detection of the presynaptic protein Synaptotagmin1 in w1118, Adar5G1 mutant, Adar5G1; ChAT>Hsc70-4, Adar5G1; ChAT>Sgt RNAi, and Adar5G1; Act5c>Hsc70-4 head protein extracts. Quantitation of the immunoblot data is shown below; levels of Synaptotagmin 1 compared to tubulin in each of the different head protein extracts. n ≤ 3. g Immunoblot to detect ref(2)p, the Drosophila p62 autophagy protein, in total head proteins of Adar5G1 mutant, w1118 wild type, Adar5G1; ChAT>Hsc70-4, and Adar5G1; ChAT>Sgt RNAi flies. n ≤ 3. h Immunoblot to detect Hsc70-4 protein in total head protein extracts of w1118 wild type, Adar5G1 mutant, Adar5G1; ChAT>Hsc70-4, and Adar5G1; ChAT>Sgt RNAi flies and Adar5G1; Act5c>Hsc70-4. n = 3. i qPCR of Hsc70-4 from w1118 wild type and Adar5G1 heads showing that Hsc70-4 is significantly decreased in Adar5G1 heads. n = 6. p values in a, e, g, and h were calculated by a one-way ANOVA followed by Tukey’s test. The significance of differences between variables was described based on p values: *p value < 0.05, **p value < 0.005, ***p value < 0.001, and n.s (not significant). Error bars: SEM (standard error of mean for biological replicates). p values in h were calculated by Student’s t test. Source data values are included in Additional file 6