Figure 1.
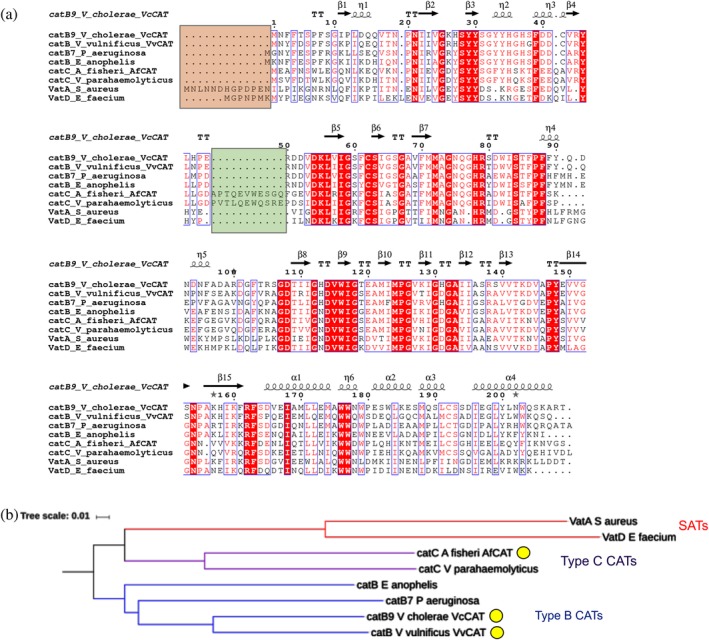
Pair‐wise sequence alignment and phylogenetic tree of CAT enzymes. (a) Sequence alignment comparing Vibrio proteins in this study to other known Type B and C CATs and SATs. The orange box highlights the differences in N‐terminus of SATs compared with Type B and C CATs. The green box highlights the alpha helical insertion of Type C CAT proteins not found in other CATs or SATs. (b) Phylogenetic tree of Type B and C CATs and SATs. Red nodes correspond to SATs, purple nodes correspond to Type C CATs, and blue nodes correspond to Type B CATs. The same sequences were used for the sequence alignment and phylogenetic tree. CATs discussed in this work are highlighted in a yellow circle. Clustal Omega, ESPRIPT, iTOL, and Microsoft PowerPoint were used to create the figure images (see the Materials and Methods section for more details)
