Figure 3 |. Mislocalization of ANCL mutants of CSPα leads to deficiencies in SNAP-25 levels and SNARE-complex assembly in neurons.
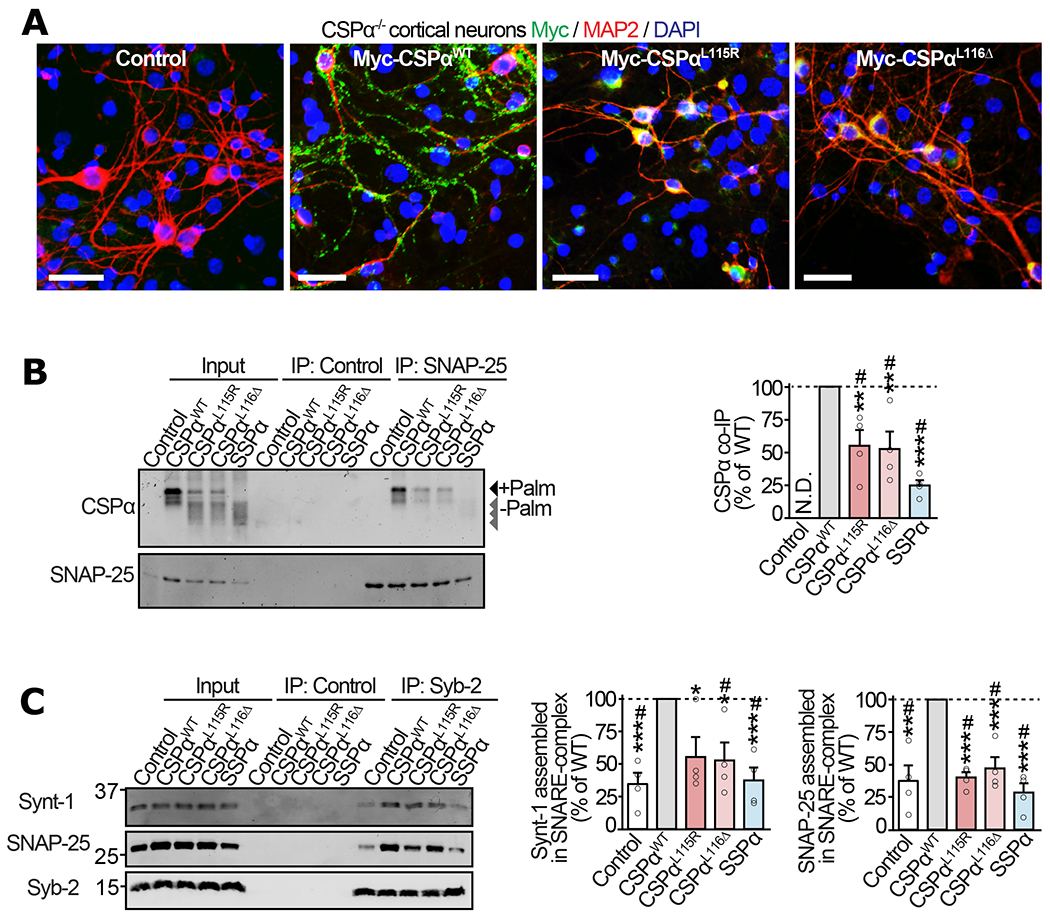
Primary cortical neurons from neonatal CSPα−/− mice were infected with lentiviruses expressing myc-tagged CSPαWT, CSPαL115R, CSPαL116Δ, SSPα, or empty lentivirus (Control) on 7 days in vitro (DIV) and examined on 17 DIV. (a) Localization of CSPα variants visualized by immunofluorescence against myc (green), somatodendritic marker MAP2 (red), and nuclear stain DAPI (blue); bar = 50 μm. (b) Interaction of CSPα variants with SNAP-25 was measured via co-immunoprecipitation (co-IP) assay. SNAP-25 IP was followed by quantitative immunoblotting of the co-IP’d CSPα (top blot), and normalized to the IP’d SNAP-25 (bottom blot). Control = sham IP without IgG. N.D. = not detected. (c) Synaptic SNARE-complex assembly was measured by synaptobrevin-2 (Syb-2) IP followed by quantitative immunoblotting of co-IP’d SNARE-complex partners syntaxin-1 (Synt-1) and SNAP-25, each normalized to the IP’d Syb-2. Control = sham IP with plain rabbit serum. Data are from (a) representative of n=3 transductions, (b) n=4 SNAP-25 immunoprecipitations, and (c) n=4 Syb-2 immunoprecipitations. In (b-c) data represent means ± SEM. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001 by two-tailed Student’s t-test. #P<0.05 by Mann-Whitney-Wilcoxon U test. Uncropped blots are shown in Source Data Fig 3.
