Figure 1.
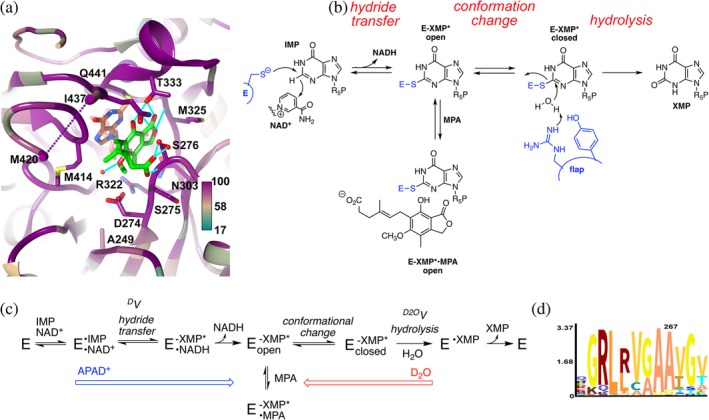
The inhibition of inosine 5′‐monophosphate dehydrogenase (IMPDH) by mycophenolic acid (MPA). (a) Structure of the MPA binding site in Cricetulus griseus (CgIMPDH2) colored by conservation (https://apps.spi-global.com/eProofing/VerifyTokenandAuthenticate.aspx?token=oIM5AEXhrAJLPSPI_PlusGnzGemOw&ChapterOrArticleOrBook=Article 27). The alignment included three MPA‐resistant IMPDHs (PbIMPDHA, PbIMPDHB, and ScIMPDH2) and seven MPA‐sensitive IMPDHs (AnIMPDH, CaIMPDH, PrIMPDHA, PrIMPDHB, ScIMPDH3, ScIMPDH4, and CgIMPDH2; alignment in Supporting Information). MPA, green; E‐XMP*, coral; K+, purple ball; residues within 4 Å of MPA and IMP are shown in stick. Ala249 is analogous to Ala267 in PbIMPDHA. (b). Mechanism of the IMPDH reaction. MPA traps E‐XMP* by binding in the vacant cofactor site after NADH dissociates, preventing the conformational change that brings the flap into the cofactor site for hydrolysis of E‐XMP*. (c). Kinetic mechanism of the IMPDH reaction. APAD+ has a lower affinity and higher redox potential than NAD+ (−0.258 V vs. −0.320),12 which will push the reaction toward E‐XMP*. D2O will make the hydrolysis step more rate limiting, which can increase the accumulation of E‐XMP*. (d). Logo graphic of conservation of sequence around position 267 in 20,000 fungal IMPDHs
