Abstract
Background
Pretreatment HIV drug resistance (HIVDR) to NNRTIs has consistently increased in Mexico City during the last decade.
Objectives
To infer the HIV genetic transmission network in Mexico City to describe the dynamics of the local HIV epidemic and spread of HIVDR.
Patients and methods
HIV pol sequences were obtained by next-generation sequencing from 2447 individuals before initiation of ART at the largest HIV clinic in Mexico City (April 2016 to June 2018). Pretreatment HIVDR was estimated using the Stanford algorithm at a Sanger-like threshold (≥20%). Genetic networks were inferred with HIV-TRACE, establishing putative transmission links with genetic distances <1.5%. We examined demographic associations among linked individuals with shared drug resistance mutations (DRMs) using a ≥ 2% threshold to include low-frequency variants.
Results
Pretreatment HIVDR reached 14.8% (95% CI 13.4%–16.2%) in the cohort overall and 9.6% (8.5%–10.8%) to NNRTIs. Putative links with at least one other sequence were found for 963/2447 (39%) sequences, forming 326 clusters (2–20 individuals). The inferred network was assortative by age and municipality (P < 0.001). Clustering individuals were younger [adjusted OR (aOR) per year = 0.96, 95% CI 0.95–0.97, P < 0.001] and less likely to include women (aOR = 0.46, 95% CI 0.28–0.75, P = 0.002). Among clustering individuals, 175/963 (18%) shared DRMs (involving 66 clusters), of which 66/175 (38%) shared K103N/S (24 clusters). Eight municipalities (out of 75) harboured 65% of persons sharing DRMs. Among all persons sharing DRMs, those sharing K103N were younger (aOR = 0.93, 95% CI 0.88–0.98, P = 0.003).
Conclusions
Our analyses suggest age- and geographically associated transmission of DRMs within the HIV genetic network in Mexico City, warranting continuous monitoring and focused interventions.
Introduction
Pretreatment HIV drug resistance (HIVDR) to first-generation NNRTIs has consistently increased in different regions across Mexico during the last decade.1 A nationally representative survey performed in 2015 showed efavirenz/nevirapine pretreatment drug resistance prevalence approaching 10%,2,3 a threshold defined by the WHO at which to recommend urgent public health action.4,5 A more recent regionally representative survey showed a similar NNRTI pretreatment HIVDR level in the central subregion including the Mexico City metropolitan area.6 This rate of HIVDR is especially relevant in Mexico (and many other Latin American countries), as the preferred first-line ART regimens were efavirenz based until modification of national ART guidelines in late 2018.7 The Mexico City metropolitan area (including Mexico City and some municipalities of the neighbouring Mexico State) includes over 15% of the nearly 250 000 persons estimated to be living with HIV (PLWH) in Mexico8,9 and is a major hub for viral dissemination within the country.10 The Condesa Clinic in Mexico City is the largest HIV care provider in Mexico, with more than 14 000 active clients (of an estimated 37 000 PLWH in Mexico City).8 The Condesa Clinic is also a key HIV detection centre, performing ∼35 000 HIV tests, and diagnosing ∼4000 new infections annually (according to the Condesa Information System), which represent over a quarter of all HIV diagnoses in Mexico and ∼70%–80% of all annual diagnoses in Mexico City metropolitan area.8 Due to its size and HIV testing volume, the Condesa Clinic is an ideal site for molecular surveillance in Mexico. Combining HIV molecular data and epidemiological information might help not only to understand HIVDR transmission dynamics locally, but also to design more efficient, cluster-informed, early detection and prevention strategies.
Genetic sequence data have been increasingly used to identify clusters of transmission for rapidly evolving pathogens, such as HIV. While HIV spreads mainly along sexual contact networks,11–13 the relatedness of HIV sequences can be used to infer the approximate transmission network.14–16 These kinds of molecular epidemiology analyses have provided insights into transmission of drug-resistant virus,17 presence of subepidemics with particular risks18 and contributions of acutely infected persons,19,20 late presenters,21 undiagnosed infections22 and cross-border/cross-community networks to local epidemics.16,23–26 In this study, we aimed to apply genetic distance-based methods to infer the local HIV genetic network in the Mexico City metropolitan area and assess the spread of drug resistance mutations (DRMs) within the network.
Patients and methods
Study population
Study enrolment took place at the Condesa Clinic’s two locations (Cuauhtémoc and Iztapalapa) in Mexico City between April 2016 and September 2018. Approximately half of all individuals testing positive during the enrolment period were offered the option to participate in this study. After providing written informed consent, blood samples were obtained and processed at a reference HIV genotyping laboratory. The study was revised and approved by the Institutional Review Board of the National Institute of Respiratory Diseases (Code: E12-17). From a total of 2496 participants enrolled, HIV sequences were successfully obtained for 2447 (98%). All individuals had plasma viral load >1000 copies/mL. Basic sociodemographic data (age, sex, state and municipality of residence) were collected at the counselling service of the clinic.
HIV sequencing
Near full-length genome HIV sequences (HXB2 positions 538–9642) were generated for all newly diagnosed individuals at Condesa participating in the study, using in-house-developed amplification protocols and deep sequencing.2 For this study, only the pol gene region was aligned and analysed. Briefly, purified viral RNA from 1 mL of plasma (QIAamp Viral RNA Mini Kit; QIAGEN, Hilden, Germany) was reverse transcribed using Superscript III Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA) and primer 1.R3.B3R 5′-ACTACTTGAAGCACTCAAGGCAAGCTTTATTG-3′ (HXB2 positions: 9611–9642) at 50°C for 1 h and 55°C for 1 h. A long-range single-round PCR was then performed with primers 1.U5.B1F 5′-CCTTGAGTGCTTCAAGTAGTGTGTGCCCGTCTGT-3′ (538–571) and 1.R3.B3R, using Expand Long Template Enzyme Mix (Roche, Basel, Switzerland) with the following conditions: 94°C for 4 min; 15 cycles of 94°C for 15 s, 68°C for 30 s and 68°C for 9.2 min; 23 cycles of 94°C for 15 s, 65°C for 30 s and 68°C for 9.2 min; and 68°C for 10 min. Genetic libraries for sequencing were generated with the Nextera XT DNA Library Prep kit (Illumina, San Diego, CA, USA), according to the manufacturer’s protocols, and sequenced using the MiSeq platform (Illumina). HIV pol sequences were then assembled using HyDRA (Public Health Agency of Canada),27,28 and consensus sequences with a Sanger-like threshold to detect genetic variants (≥20%)29 were obtained for each participant for HIVDR analysis. Consensus sequences were quality filtered, and a local BLAST was performed to exclude sequences <0.1% different from historical sequences with the same associated birth date. HIVDR prevalence was estimated using the Stanford algorithm (v8.4) with the HIVdb tool,30,31 defining viruses with a Stanford score ≥15 to efavirenz, nevirapine, any NRTI, lopinavir, atazanavir, darunavir, raltegravir, elvitegravir or dolutegravir as resistant, as recommended by WHO standardized protocols.32 Analysis of shared DRMs was performed considering variants from the 2009 surveillance DRM list.33 DRM sharing was defined as the presence of the same DRM in two linked sequences at a ≥2% sensitivity threshold, in order to include both low-frequency variants (2%–19%) and variants at Sanger-like frequency (≥20%).
Genetic network inference
The HIV genetic network was inferred with HIV-TRACE,34 establishing putative transmission links between PLWH whose HIV sequences had a genetic distance of <1.5%. Inferred links were then resolved into clusters for further analysis. The genetic network was reconstructed after removing all of the major DRMs33 from the sequences so that they would not impact the genetic distance comparison, but the resulting network was unchanged. Newman’s assortativity coefficients for age and location of residence were estimated using the R package igraph. Null distributions to assess assortativity significance were obtained with the R sample function to create a random distribution with 1000 iterations.35 Geospatial dispersal was determined by calculating the average distance between centroids of the municipalities of residence of genetically linked individuals.
Discrete diffusion model
To reduce the computation burden, sequences were subsampled to best approximate the epidemic dynamics.36 Briefly, the study sequences were complemented with all location-annotated, publicly available HIV subtype B pol sequences. A phylogenetic tree was inferred using FastTree237 under the GTR + Γ evolutionary model. Strongly supported clades including only sequences from Mexico City’s metropolitan area and having Shimodaira–Hasegawa local support ≥0.9 were identified.38 Next, well-supported monophyletic clades including only sequences sampled from the same municipality were identified. From each of these clades, one sequence was randomly selected for inclusion in downstream analyses.39
Phylogeographic inference was performed using the discrete diffusion model40 implemented in BEAST 1.10.4.41 Migration patterns were reconstructed utilizing the asymmetric diffusion model,40,42 using individual municipalities as location traits. To identify the subset of migration rates most informative to reconstruct the dispersal history, we used a model averaging procedure (Bayesian stochastic search variable selection).40 Bayes factor (BF) support for all possible types of location exchanges was calculated with SpreaD3.43 BFs above 20 were considered to be strong support for a link between sampled locations.44 Estimates of the posterior probability of the expected number of migration events between all pairs of locations were computed through stochastic mapping techniques.45,46 To investigate whether support for transition rates was associated with sampling heterogeneity, analyses were repeated while randomly permuting location states between tips during Markov chain Monte Carlo (MCMC) sampling.47 Migration links with good BF support in the ‘as is’ analysis and poor BF support in the ‘tip-state-swap’ analysis were considered for further analysis. We refer to this as the adjusted BF. MCMC chains were run to ensure adequate mixing. Maximum clade credibility trees were obtained with TreeAnnotator 1.10,41 and convergence and mixing properties were inspected using Tracer 1.7.48
Statistical analysis
Available demographic metadata were explored, curated and checked for missing data and errors. Crude ORs and 95% CIs between outcome and exposure variables were estimated independently and significance of associations was assessed by χ2 tests for OR = 1. For ordered categorical variables, we obtained stratum-specific odds and tested for trends. To account for confounding and effect modification, multivariable logistic regression models were constructed to explore associations of sociodemographic variables (including age, sex, municipality of residence, cluster size and sampling year) with belonging to clusters or with sharing DRMs within the network. All analyses were performed using STATA (v15).
Results
HIV drug resistance prevalence in Mexico City metropolitan area
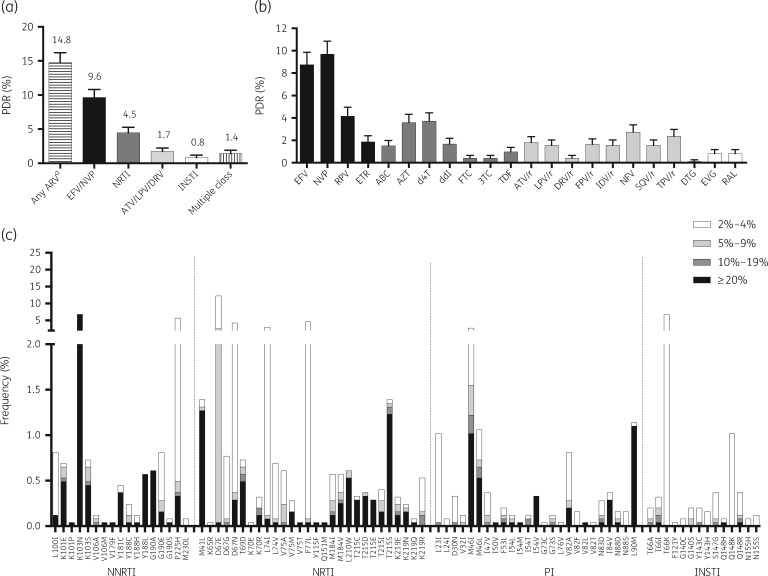
After quality filtering, considering all positions associated with HIVDR, the median read coverage was 2333 (IQR 1126–5053). At the Sanger-like threshold of ≥20% and considering WHO definitions,32 pretreatment HIVDR reached 14.8% (95% CI 13.4%–16.2%) to any antiretroviral, 9.6% (8.5%–10.8%) to NNRTIs (efavirenz, nevirapine) and 4.5% (3.6%–5.3%) to NRTIs. As expected given their less common use in first-line ART regimens, pretreatment HIVDR to boosted PIs (lopinavir, atazanavir, darunavir; 1.7%, 1.2%–2.2%) and integrase strand transfer inhibitors (INSTIs; 0.8%, 0.5%–1.2%) was low (Figure 1a). HIVDR to single drugs was highest for efavirenz (8.7%) and nevirapine (9.6%), and lowest for dolutegravir (0.1%) and darunavir (0.4%) (Figure 1b). K103N was the most frequent surveillance DRM with 6.6% frequency (7.1% K103N/S) (Figure 1c). Other commonly observed NNRTI DRMs included G190A, Y188L, Y181C and K101E (Figure 1c). Both M184V and M184I were observed mainly as low-frequency variants in approximately 0.6% of the participants. K65R was very rare and only observed as a low-frequency variant. Frequent NRTI DRMs included the thymidine analogue mutations M41L, L210W and T215S. The most frequent PI mutations included M46I/L and L90M. INSTI mutations were rare (Figure 1c). The frequencies of all DRMs included in the Stanford HIVDR Database are shown in Tables S1–S4 (available as Supplementary data at JAC Online).
Figure 1.
HIV pretreatment drug resistance (PDR) prevalence in the Mexico City metropolitan area, 2016–18. HIV drug resistance was estimated using next-generation sequencing from 2447 persons starting first-line ART, diagnosed at the Condesa Clinic. (a) HIVDR prevalence by drug class. (b) HIVDR prevalence by antiretroviral drug. Lines represent 95% CI. (c) DRM frequency at different sensitivity thresholds; only surveillance mutations are shown (Stanford HIV Drug Resistance Database). EFV, efavirenz; NVP, nevirapine; RPV, rilpivirine; ETR, etravirine; ABC, abacavir; AZT, zidovudine; d4T, stavudine; ddI, didanosine; FTC, emtricitabine; 3TC, lamivudine; TDF, tenofovir disoproxil fumarate; ATV/r, atazanavir boosted with ritonavir; LPV/r, lopinavir boosted with ritonavir; DRV/r, darunavir boosted with ritonavir; FPV/r, fosamprenavir boosted with ritonavir; IDV/r, indinavir boosted with ritonavir; NFV, nelfinavir; SQV/r, saquinavir boosted with ritonavir; TPV/r, tipranavir boosted with ritonavir; DTG, dolutegravir; EVG, elvitegravir; RAL, raltegravir; INSTI, integrase strand-transfer inhibitor. aConsidering EFV, NVP, any NRTI, ATV/r, LPV/r, DRV/r, DTG, EVG and RAL.
HIV genetic network in Mexico City metropolitan area
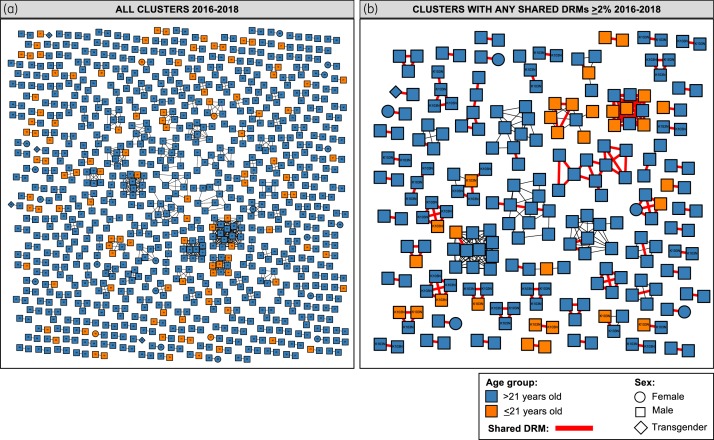
Putative links with at least one other sequence were found for 963/2447 (39%) sequences, forming 326 clusters ranging in size from 2 to 20 individuals (Figure S1). Clustering individuals were younger [adjusted OR (aOR) per 1 year increase = 0.96, 95% CI 0.95–0.97, P < 0.001], less likely to include women (aOR = 0.46, 0.28–0.75, P = 0.002) and less likely to reside outside of the central metropolitan area (aOR = 0.12, 0.03–0.51, P = 0.004) (Table 1). Persons 20–24 years old were over-represented and persons ≥40 years old were under-represented within clustering individuals (P < 0.001; Table S5). Young persons (≤21 years old) comprised 14% (139/963) of clustering individuals, belonging to 103 clusters (Figure 2a). Of all putative links (1158), 24% (278) included at least one person ≤21 years old. Of these links, 42% (117/278) included one 22 to 25 year old, 38% (106/278) one 26 to 40 year old, 6% (16/278) one person >40 years old, and 23% (63/278) another person ≤21 years old, suggesting frequent putative links between persons of diverse age groups. Persons ≤21 years old were not more prone to link with persons of their same age group compared with persons >40 years old (11.8% versus 9.5%, P = 0.6), while persons ≤30 years old were more prone to link with persons of their same age compared with persons >30 years old (47.6% versus 26.2%, P < 0.0001). Moreover, persons <25 years old were more likely to belong to larger clusters (size >2: OR = 1.3, 1.03–1.8, P = 0.003; size >9: OR = 1.7, 1.06–2.7, P = 0.02). When estimating Newman’s coefficients, the inferred transmission network was assortative by age (0.0867; P < 0.0001) and municipality of residence (0.0077; P < 0.001) (Figure S2). Considering centroids of municipalities, genetically linked individuals were more likely to live closer (13 km median observed distance versus 65 km expected distance; P < 0.01). The proportion of persons harbouring major DRMs in protease was slightly higher in clustering individuals (3.3% versus 2.0%; OR: 1.7, 1.0–2.8, P = 0.045), but significance was lost after adjusting for age, sex, year of enrolment and location of residence (Table 1).
Table 1.
Variables associated with clustering within Mexico City’s HIV genetic network
| Variables and categories | Clustering n (%) | Crude OR (95% CI) | P | Adjusted OR (95% CI)a | P |
|---|---|---|---|---|---|
| Age | — | 0.96 (0.96–0.97) | <0.001 | 0.96 (0.95–0.97) | <0.001 |
| Sex | |||||
| male (n=2322) | 938 (40.4) | 1.00 | 1.00 | ||
| female (n=105) | 21 (20.0) | 0.37 (0.23–0.60) | <0.001 | 0.46 (0.28–0.75) | 0.002 |
| transgender (n=16) | 4 (25.0) | 0.49 (0.16–1.53) | 0.211 | 0.54 (0.17–1.70) | 0.294 |
| Year of enrolment | |||||
| 2016 (n=448) | 177 (39.5) | 1.00 | 1.00 | ||
| 2017 (n=1784) | 705 (39.5) | 1.00 (0.81–1.24) | 0.997 | 1.08 (0.87–1.34) | 0.502 |
| 2018 (n=213) | 80 (37.6) | 0.92 (0.66–1.29) | 0.631 | 0.98 (0.69–1.39) | 0.903 |
| Municipality | |||||
| other CDMX (n=612) | 235 (38.4) | 1.00 | 1.00 | ||
| other Mexico State (n=397) | 162 (40.8) | 1.11 (0.85–1.43) | 0.445 | 1.06 (0.82–1.39) | 0.640 |
| Gustavo A. Madero (n=234) | 94 (40.2) | 1.08 (0.79–1.47) | 0.636 | 1.06 (0.78–1.46) | 0.695 |
| Iztacalco (n=73) | 33 (45.2) | 1.32 (0.81–2.16) | 0.260 | 1.29 (0.78–2.13) | 0.315 |
| Iztapalapa (n=238) | 100 (42.0) | 1.16 (0.86–1.58) | 0.333 | 1.14 (0.84–1.56) | 0.399 |
| Álvaro Obregón (n=108) | 37 (34.3) | 0.84 (0.54–1.29) | 0.414 | 0.81 (0.52–1.26) | 0.351 |
| Benito Juárez (n=137) | 57 (41.6) | 1.14 (0.78–1.67) | 0.487 | 1.18 (0.80–1.73) | 0.394 |
| Cuauhtémoc (n=344) | 134 (39.0) | 1.02 (0.78–1.34) | 0.866 | 1.03 (0.78–1.36) | 0.829 |
| Ecatepec (n=91) | 33 (36.3) | 0.91 (0.58–1.44) | 0.696 | 0.84 (0.53–1.34) | 0.466 |
| Nezahualcóyotl (n=91) | 40 (44.0) | 1.25 (0.81–1.96) | 0.311 | 1.22 (0.78–1.92) | 0.390 |
| other states (n=30) | 2 (6.7) | 0.11 (0.03–0.49) | <0.001 | 0.12 (0.03–0.51) | 0.004 |
| unknown (n=92) | 36 (39.1) | 1.03 (0.66–1.62) | 0.893 | 0.92 (0.58–1.47) | 0.725 |
| Major DRMs in RT (n=269) | 114 (42.4) | 1.15 (0.89–1.49) | 0.282 | 1.09 (0.84–1.42) | 0.518 |
| Major DRMs in PR (n=62) | 32 (51.6) | 1.67 (1.01–2.76) | 0.045 | 1.58 (0.93–2.68) | 0.089 |
| Major DRMs in IN (n=3) | 1 (33.3) | 0.77 (0.07–8.51) | 0.831 | 0.56 (0.05–6.54) | 0.644 |
Statistically significant P values are indicated in bold.
CDMX, Mexico City; RT, reverse transcriptase; PR, protease; IN, integrase.
Logistic regression model including age, sex, year of enrolment, municipality and presence of major DRMs in PR, RT or IN. Number of observations: 2334. Data on year of enrolment are missing for 2 individuals. Data on age are missing for 11 individuals. Data on sex are missing for 4 individuals.
Figure 2.
Contribution of young persons in Mexico City’s HIV genetic network, 2016–18. Clusters are coloured by age group. (a) All clusters are shown; age at enrolment is included within each node. (b) Only clusters with putative sequence pairs sharing DRMs at ≥2% threshold are shown; shared K103N is identified. Node shapes denote gender. All edges represent a genetic distance of <1.5% separating nodes. Red edges denote putative sequence pairs sharing DRMs. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
HIVDR transmission within the HIV genetic network
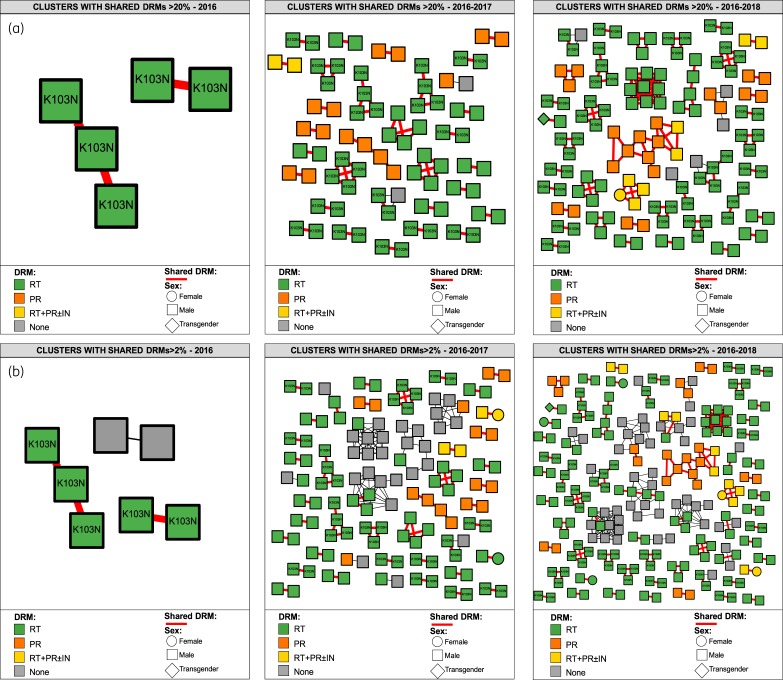
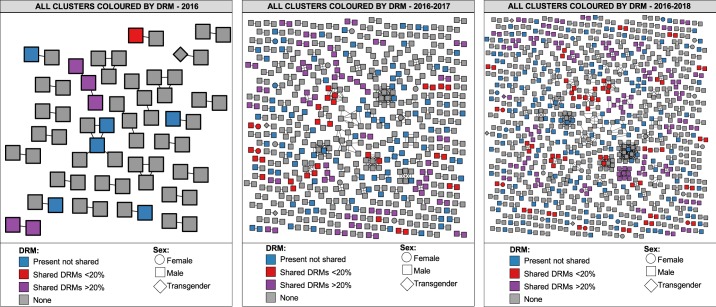
Among clustering individuals, 175/963 (18%) shared DRMs involving 66 clusters. Of these clusters, 25 included persons sharing only low-frequency DRMs. Among all individuals sharing DRMs, 66/175 (38%) shared K103N/S involving 24 clusters (Figures 3a and 3b). Other frequently shared DRMs included: reverse transcriptase (RT) Y188L (11/175), RT T215S/C (24/175) and protease M46I (19/175), mostly at ≥20% within-host frequency; and RT D67N/G/E (56/175) and RT P225H (16/175) as low-frequency variants. Persons sharing DRMs were more frequently enrolled after 2016 (2017 aOR = 1.7, 1.0–2.7, P = 0.04; 2018 aOR = 2.4, 1.2–5.0, P = 0.02) (Table 2). Indeed, the odds of sharing DRMs increased with time (0.15, 0.23 and 0.31 for 2016, 2017 and 2018, respectively; test for trend P = 0.04), observing 12%, 19% and 24% of persons sharing DRMs (among clustering individuals) participating in 3, 46 and 66 clusters, in 2016, 2017 and 2018, respectively (Figure 4). Young persons ≤21 years old participated in 19 out of 66 clusters including persons sharing DRMs (Figure 2b). As expected, low-frequency DRMs were less commonly observed in all members of the cluster than when using the ≥20% threshold (Figures 3 and 4). Analysing the geographical distribution of persons within the network, eight municipalities (out of 75)—six in Mexico City (Gustavo A. Madero, Cuauhtémoc, Iztapalapa, Álvaro Obregón, Benito Juárez and Iztacalco) and two in Mexico State (Ecatepec and Nezahualcóyotl)—encompassed the places of residence of 65% of all persons sharing DRMs (Figure 3c). Of note, the Gustavo A. Madero municipality had 3.2 times the odds (95% CI 1.7–5.8, P < 0.001) of including persons sharing DRMs than other municipalities within Mexico City (Table 2). A similar trend was also strongly significant for Iztapalapa (aOR = 2.6, 1.4–4.9, P = 0.002) and Ecatepec (aOR = 3.5, 1.5–8.1, P = 0.004) (Table 2).
Figure 3.
Cluster growth and geographic residence of persons sharing DRMs. (a) Clusters with shared DRMs at ≥20% sensitivity threshold. (b) Clusters with shared DRMs at ≥2% threshold. Nodes are coloured by HIV gene. Node shapes denote gender. All edges represent a genetic distance of <1.5% separating nodes. Red edges denote putative sequence pairs sharing DRMs. Sequences sharing K103N are shown. Annual growth of the network is shown from left to right. (c) Geographic residence of individuals sharing DRMs. Maps of Mexico City metropolitan area divided into municipalities are shown. Eight municipalities are shown, which accounted for 65% of persons sharing DRMs. IN, integrase. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Table 2.
Variables associated with sharing DRMs among all persons within clusters in Mexico City’s HIV genetic network
| Variables and categories | Sharing DRMs n (%) | Crude OR (95% CI) | P | Adjusted OR (95% CI)a | P |
|---|---|---|---|---|---|
| Age | — | 1.00 (0.98–1.02) | 0.779 | 1.00 (0.98–1.02) | 0.781 |
| Sex | |||||
| male (n=938) | 170 (18.1) | 1.00 | 1.00 | ||
| female (n=21) | 4 (19.1) | 1.06 (0.35–3.20) | 0.914 | 0.96 (0.31–2.98) | 0.948 |
| Transgender (n=4) | 1 (25.0) | 1.51 (0.16–14.59) | 0.722 | 1.92 (0.19–19.92) | 0.584 |
| Cluster size | — | 0.98 (0.94–1.03) | 0.408 | 0.99 (0.94–1.04) | 0.632 |
| Year of enrolment | |||||
| 2016 (n=177) | 22 (12.4) | 1.00 | 1.00 | ||
| 2017 (n=705) | 134 (19.0) | 1.65 (1.02–2.69) | 0.040 | 1.66 (1.02–2.72) | 0.043 |
| 2018 (n=80) | 19 (23.8) | 2.19 (1.10–4.38) | 0.022 | 2.40 (1.16–4.96) | 0.018 |
| Municipality | |||||
| other CDMX (n=235) | 26 (11.1) | 1.00 | 1.00 | ||
| other Mexico State (n=162) | 30 (18.5) | 1.83 (1.03–3.24) | 0.036 | 1.77 (1.00–3.14) | 0.052 |
| Gustavo A. Madero (n=94) | 26 (27.7) | 3.07 (1.65–5.73) | <0.001 | 3.16 (1.71–5.84) | <0.001 |
| Iztacalco (n=33) | 6 (18.2) | 1.79 (0.67–4.75) | 0.239 | 1.65 (0.61–4.40) | 0.321 |
| Iztapalapa (n=100) | 25 (25.0) | 2.68 (1.44–4.98) | 0.001 | 2.63 (1.43–4.86) | 0.002 |
| Álvaro Obregón (n=37) | 9 (24.3) | 2.58 (1.09–6.13) | 0.025 | 2.43 (1.03–5.77) | 0.044 |
| Benito Juárez (n=57) | 10 (17.5) | 1.71 (0.77–3.80) | 0.183 | 1.73 (0.78–3.84) | 0.180 |
| Cuauhtémoc (n=134) | 21 (15.7) | 1.49 (0.80–2.78) | 0.202 | 1.51 (0.81–2.81) | 0.196 |
| Ecatepec (n=33) | 10 (30.3) | 3.49 (1.47–8.29) | 0.003 | 3.45 (1.47–8.09) | 0.004 |
| Nezahualcóyotl (n=40) | 6 (15.0) | 1.42 (0.54–3.71) | 0.474 | 1.37 (0.52–3.59) | 0.524 |
| other states (n=2) | 0 (0) | — | — | — | — |
| unknown (n=36) | 6 (16.7) | 1.61 (0.61–4.24) | 0.333 | 1.09 (0.38–3.18) | 0.869 |
Statistically significant P values are indicated in bold.
CDMX, Mexico City.
Logistic regression model including age, sex, cluster size, year of enrolment and municipality. Number of observations: 958. Data on year of enrolment are missing for one individual. Data on age are missing for three individuals.
Figure 4.
Linked drug resistance and growth of the Mexico City HIV genetic network 2016–18. Clusters are coloured by the presence of shared DRMs at ≥20% and 2%–19% sensitivity thresholds. All edges represent a genetic distance of <1.5% separating nodes. Annual growth of the network is shown from left to right. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
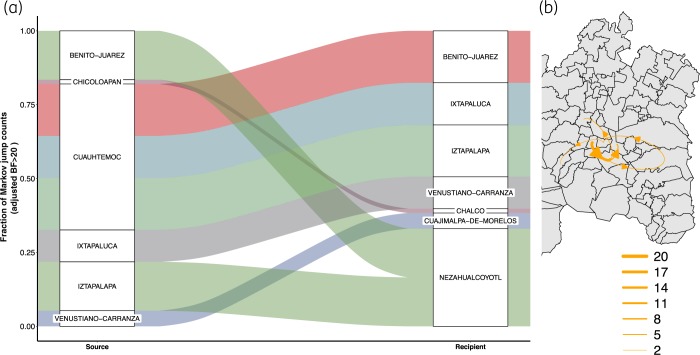
In our phylogeographic diffusion model, the municipalities of Cuauhtémoc, Iztapalapa, Benito Juárez (in Mexico City) and Ixtapaluca (in Mexico State) were the most strongly supported (BF >20) sources of HIV, while Nezahualcóyotl, Ixtapaluca (in Mexico State), Iztapalapa, Benito Juárez and Venustiano Carranza (in Mexico City) were commonly recipients (Figure 5a). This analysis identifies a ‘core HIV diffusion area’ across the northern part of Mexico City and the neighbouring municipalities to the east (Figure 5b). Interestingly, Gustavo A. Madero was not included in any strongly supported transition, suggesting local persistence and spread of drug-resistant viruses.
Figure 5.
Lineage dispersal events between municipalities in the Mexico City metropolitan area. (a) Sankey plot showing the proportion of transition events from each source municipality (left) toward the recipient municipality (right). Only strongly supported transitions (adjusted BF >20) are shown and are coloured by source. The size of the boxes is proportional to the number of transitions observed. (b) Number of lineage dispersal events between municipalities. The thickness of the arrows corresponds to the average number of strongly supported, inferred migration events between locations (BF >20). This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Among all persons sharing DRMs (n = 175), those sharing K103N were significantly younger (aOR per 1 year increase: 0.93, 0.88–0.98, P = 0.003), were less prone to belong to larger clusters (aOR per cluster size unit increase: 0.66, 0.53–0.82, P < 0.001) and were less frequent after 2016 (2017 aOR = 0.25, 0.08–0.80, P = 0.016) (Table 3).
Table 3.
Variables associated with sharing K103N among all persons sharing DRMs within Mexico City’s HIV genetic network
| Variables and categories | Sharing K103N, n (%) | Crude OR (95% CI) | P | Adjusted OR (95% CI)a | P |
|---|---|---|---|---|---|
| Age | — | 0.97 (0.93–1.00) | 0.068 | 0.93 (0.88–0.98) | 0.003 |
| Cluster size | — | 0.80 (0.71–0.89) | <0.001 | 0.66 (0.53–0.82) | <0.001 |
| Year of enrolment | |||||
| 2016 (n=22) | 12 (54.6) | 1.00 | 1.00 | ||
| 2017 (n=134) | 44 (32.8) | 0.41 (0.16–1.03) | 0.050 | 0.25 (0.08–0.80) | 0.016 |
| 2018 (n=19) | 10 (52.6) | 0.93 (0.27–3.22) | 0.904 | 0.46 (0.08–1.70) | 0.312 |
| Municipality | |||||
| other CDMX (n=26) | 10 (38.5) | 1.00 | 1.00 | ||
| other Mexico State (n=30) | 12 (40.0) | 1.07 (0.36–3.16) | 0.907 | 0.75 (0.23–2.48) | 0.635 |
| Gustavo A. Madero (n=26) | 6 (23.1) | 0.48 (0.14–1.65) | 0.234 | 0.37 (0.10–1.38) | 0.138 |
| Iztacalco (n=6) | 1 (16.7) | 0.32 (0.03–3.40) | 0.319 | 0.16 (0.01–1.91) | 0.147 |
| Iztapalapa (n=25) | 13 (52.0) | 1.73 (0.56–5.39) | 0.336 | 1.75 (0.50–6.15) | 0.383 |
| Álvaro Obregón (n=9) | 4 (44.4) | 1.28 (0.27–6.08) | 0.756 | 0.76 (0.15–3.97) | 0.748 |
| Benito Juárez (n=10) | 3 (30.0) | 0.69 (0.14–3.37) | 0.641 | 0.48 (0.08–2.79) | 0.419 |
| Cuauhtémoc (n=21) | 8 (38.1) | 0.98 (0.30–3.26) | 0.980 | 1.99 (0.46–8.54) | 0.353 |
| Ecatepec (n=10) | 3 (30.0) | 0.69 (0.14–3.37) | 0.641 | 0.54 (0.10–3.06) | 0.490 |
| Nezahualcóyotl (n=6) | 3 (50.0) | 1.60 (0.26–9.88) | 0.610 | 1.76 (0.25–12.54) | 0.572 |
| other states (n=0) | 0 (0) | — | — | — | — |
| unknown (n=6) | 3 (50.0) | 1.60 (0.26–9.88) | 0.610 | 0.94 (0.89–9.81) | 0.956 |
Statistically significant P values are indicated in bold.
CDMX, Mexico City.
Logistic regression model including age, cluster size, year of enrolment and municipality. Number of observations: 174. All participants sharing K103N were male. Data on age are missing for one individual.
Taken together, these analyses suggest age- and geographically dependent transmission of DRMs within the HIV genetic network in Mexico City. Overall, transmission of DRMs was concentrated in specific municipalities, mainly Gustavo A. Madero and Iztapalapa in Mexico City and Ecatepec in Mexico State, and increased with time.
Discussion
HIV pretreatment drug resistance to efavirenz/nevirapine in ART initiators in the largest HIV clinic in Mexico City was similar to that observed at the national representative survey in Mexico,6 and approached the 10% threshold recommended by the WHO to take immediate public health action. Recent meta-analyses and global reports evidence increasing HIVDR trends to NNRTIs in most of the developing world,49 reaching worrying levels in some Latin American and African countries.3 Leveraging on a policy window provided by administrative changes in the Mexican Government, Mexican ART Guidelines were modified, recommending dolutegravir- and bictegravir-based single-tablet regimens as preferred options, and moving efavirenz- and lower genetic barrier INSTI-based regimens to the ‘alternative’ category.50 This transition period warrants continuous HIVDR surveillance and a better understanding of HIVDR transmission dynamics, especially when no specific additional programmes to strengthen adherence and viral load monitoring have been enforced, and drug stock-outs may occur due to purchase and distribution adjustments. This is relevant given the remarkable increase in NNRTI-transmitted HIVDR during the last decade in Mexico City from 2% to 10%, which can in part be linked to programmatic weaknesses including suboptimal viral load monitoring, but also to epidemiological factors including low education, marginalization, discrimination and poverty.1,6
In this work, HIVDR was defined at the 20% threshold to make our results comparable to other studies performed with Sanger sequencing.1,3,6 Recent evidence suggests that using this threshold provides the most similar consensus sequences to Sanger.51 For HIV network analyses, we used a 2% threshold to be able to assess transmission of low frequency (2%–19%) variants, with 2% being a methodological detection limit associated with our experimental design. Given a read coverage IQR for all positions associated with HIVDR of 1126–5053 (median 2333), and considering that all participants had plasma viral loads >1000 copies/mL, our acceptance criteria to detect DRMs at 2% within-host frequency threshold varied between 23 and 101 high-quality reads (median 47), giving us high confidence that the mutations observed were not spurious.
Propagation of K103N within the network is noteworthy, and of high relevance not only to Mexico but also to many other resource-limited countries that still use efavirenz-based first-line ART regimens without availability of baseline HIV genotyping and weak viral load monitoring systems. The lack of fitness costs associated with this mutation52 in addition to the low-genetic barrier of efavirenz and its wide use as part of first-line ART regimens have resulted in worryingly high K103N prevalence in several countries.3 These observations, together with our current study, support the hypothesis of an endemic self-sustained reservoir of specific DRMs that persists in ART-naive populations mostly through onward transmission.52 This lesson needs to be taken into account in countries migrating to INSTI-based regimens, especially if low-genetic barrier drugs are to be used, considering the predicted impact of different DRMs on viral fitness and drug resistance in the context of possible interactions between DRMs in RT and integrase.53,54 In our study, local transmission of HIVDR variants was notable for the Gustavo A. Madero municipality, which showed the highest frequency of persons sharing DRMs, but was not included among the strongly supported transition events in the phylogeographic diffusion model, suggesting an important role of locally sustained transmission. However, strongly supported transition events across municipalities, including Benito Juárez–Nezahualcóyotl, Cuauhtémoc–Benito Juárez, Cuauhtémoc–Iztapalapa and Iztapalapa–Nezahualcóyotl, could partly explain HIVDR spread, mostly in the northern area of Mexico City, which was identified as the geographic epicentre of Mexico City’s epidemic.
Sexual transmission of low-frequency DRMs55–57 and their role in ART failure2,58–60 remains a controversial issue. Here, sharing of specific DRMs at low within-host frequency (2%–19%), mainly RT D67N/G/E and P225H, between putative transmission pairs was observed relatively frequently (25 out of 66 clusters including persons sharing DRMs). However, sharing of low-frequency DRMs was not consistent among all persons belonging to the cluster. As most participants in the present study were detected in the chronic phase of HIV infection, it is not possible to distinguish actual transmission versus de novo selection of these low-frequency mutations, as suggested previously.56 Moreover, consistent with previous observations,52 DRMs with low transmission fitness due to high replicative impairment such as M184V/I and K65R were not shared within the Mexican network, and were observed mostly at low within-host frequencies in the study population. This suggests different transmission, persistence, emergence and reversion mechanisms for different HIVDR variants.
Interestingly, DRM sharing patterns within Mexico City’s HIV network were closely associated with both age and geographical place of residence. The presence of at least three municipalities with strong evidence of higher odds of harbouring putative transmission chains sharing drug-resistant variants is noteworthy. Indeed, Iztapalapa, Gustavo A. Madero and Álvaro Obregón in Mexico City and Ecatepec (together with Nezahualcóyotl) in Mexico State are the most highly populated municipalities in the metropolitan area.61 Gustavo A. Madero and Iztapalapa also encompassed a high number of PLWH included in the study cohort. Whether this observation simply reflects higher sampling density of local networks or a real unequal geographical distribution needs further follow-up and observation. It is important to note that there are no municipality-specific clinics in the area, and ART is provided by centralized care centres. Thus, we are unable to look at municipality-specific treatment factors that could explain our findings. Our observations suggest an opportunity for social research to study specific socioeconomic, cultural or geographic determinants that could influence HIVDR spread, and enrich our understanding of the local epidemic. Moreover, the higher odds of sharing K103N between younger persons and in smaller clusters also suggests important groups for intervention. Whether the decreasing trend of K103N sharing observed in the network is due to enrolment bias or to a real epidemiological effect remains to be explored.
Our study has limitations that need to be acknowledged. Even though the Condesa Clinic diagnoses a large proportion of all the new cases of HIV in Mexico City annually, inclusion of other clinics in Mexico City is warranted to increase network density and representativeness. Our study may also suffer from enrolment bias, as only ∼50% of persons diagnosed at the Condesa Clinic were enrolled due to logistical complications such as materials stock-outs, laboratory closures due to maintenance and the availability of counsellors and project personnel, who were mainly present during the morning shifts. Additionally, as previously suggested,62,63 incomplete sampling may increase the chance of linking individuals who are not direct transmission partners in the network and thus bias age assortativity analyses. Also, in this study, the contribution to the network of important risk groups was not assessed, including persons failing ART, adolescents under 18 years and migrants. Future molecular surveillance in the region will greatly benefit from more detailed metadata, including specific sexual practices, venues and apps for finding sexual partners, use of recreational drugs, etc.
In conclusion, DRMs are frequently transmitted in Mexico City’s metropolitan area, predominantly among recently diagnosed young men in a geographically and age-assortative network. Sharing of specific DRMs was common, especially K103N/S, revealing the potential for spread of pretreatment HIVDR. We also identified specific geographical regions and risk groups amenable to immediate focused intervention.
Supplementary Material
Acknowledgements
We thank all participants for their involvement and generosity.
Funding
This work was supported by grants from the Mexican Government (Comisión de Equidad y Género de las Legislaturas LX-LXI y Comisión de Igualdad de Género de la Legislatura LXII de la H. Cámara de Diputados de la República Mexicana), received by G.R.-T.; and Consejo Nacional de Ciencia y Tecnología (CONACyT SALUD-2017-01-289725), received by S.A.-R. A.C. and S.R.M. acknowledge funding from the University of California San Diego Centre for AIDS Research (CFAR), an NIH-funded programme. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Transparency declarations
None to declare.
References
- 1. Garcia-Morales C, Tapia-Trejo D, Quiroz-Morales VS. et al. HIV pretreatment drug resistance trends in three geographic areas of Mexico. J Antimicrob Chemother 2017; 72: 3149–58. [DOI] [PubMed] [Google Scholar]
- 2. Avila-Rios S, Garcia-Morales C, Matias-Florentino M. et al. Pretreatment HIV-drug resistance in Mexico and its impact on the effectiveness of first-line antiretroviral therapy: a nationally representative 2015 WHO survey. Lancet HIV 2016; 3: e579–91. [DOI] [PubMed] [Google Scholar]
- 3.WHO. HIV Drug Resistance Report 2019. https://www.who.int/hiv/pub/drugresistance/hivdr-report-2019/en/.
- 4.WHO. Global Action Plan on HIV Drug Resistance 2017. –2021. http://www.who.int/hiv/pub/drugresistance/hivdr-action-plan-2017-2021/en/.
- 5.WHO. Guidelines on the Public Health Response to Pretreatment HIV Drug Resistance. 2017. http://www.who.int/hiv/pub/guidelines/hivdr-guidelines-2017/en/.
- 6. Avila-Rios S, Garcia-Morales C, Valenzuela-Lara M. et al. HIV-1 drug resistance before initiation or re-initiation of first-line ART in eight regions of Mexico: a sub-nationally representative survey. J Antimicrob Chemother 2019; 74: 1044–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.CENSIDA/Secretaría de Salud. Guía de Manejo Antirretroviral de las Personas con VIH. Novena Edición. Gobierno de México. 2018. https://www.gob.mx/censida/articulos/guia-de-manejo-antirretroviral-de-las-personas-con-vih-89591? idiom=es.
- 8.CENSIDA. Epidemiología/Registro Nacional de Casos de VIH y SIDA. Cierre. 2018. https://www.gob.mx/censida/documentos/epidemiologia-registro-nacional-de-casos-de-sida.
- 9.CENSIDA. Informe Nacional del Monitoreo de Compromisos y Objetivos Ampliados para Poner Fin al Sida (Informe GAM). Mexico. 2018. https://www.gob.mx/censida/documentos/informe-nacional-del-monitoreo-de-compromisos-y-objetivos-ampliados-para-poner-fin-al-sida-informe-gam-mexico-2018? idiom=es.
- 10. Chaillon A, Avila-Rios S, Dennis A. et al. Phylogeographic analysis suggests gravity model of HIV transmission in Mexico. Conference on Retroviruses and Opportunistic Infections 2018, Boston, MA, USA, 2018. Poster 954.
- 11. Klovdahl AS. Social networks and the spread of infectious diseases: the AIDS example. Soc Sci Med 1985; 21: 1203–16. [DOI] [PubMed] [Google Scholar]
- 12. Rothenberg RB, Potterat JJ, Woodhouse DE. et al. Social network dynamics and HIV transmission. AIDS 1998; 12: 1529–36. [DOI] [PubMed] [Google Scholar]
- 13. Woodhouse DE, Rothenberg RB, Potterat JJ. et al. Mapping a social network of heterosexuals at high risk for HIV infection. AIDS 1994; 8: 1331–6. [DOI] [PubMed] [Google Scholar]
- 14. Leigh Brown AJ, Lycett SJ, Weinert L. et al. Transmission network parameters estimated from HIV sequences for a nationwide epidemic. J Infect Dis 2011; 204: 1463–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rothenberg R. HIV transmission networks. Curr Opin HIV AIDS 2009; 4: 260–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wertheim JO, Leigh Brown AJ, Hepler NL. et al. The global transmission network of HIV-1. J Infect Dis 2014; 209: 304–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Panichsillapakit T, Smith DM, Wertheim JO. et al. Prevalence of transmitted HIV drug resistance among recently infected persons in San Diego, CA 1996–2013. J Acquir Immune Defic Syndr 2016; 71: 228–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hue S, Clewley JP, Cane PA. et al. HIV-1 pol gene variation is sufficient for reconstruction of transmissions in the era of antiretroviral therapy. AIDS 2004; 18: 719–28. [DOI] [PubMed] [Google Scholar]
- 19. Mehta SR, Murrell B, Anderson CM. et al. Using HIV sequence and epidemiologic data to assess the effect of self-referral testing for acute HIV infection on incident diagnoses in San Diego, California. Clin Infect Dis 2016; 63: 101–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Volz EM, Koopman JS, Ward MJ. et al. Simple epidemiological dynamics explain phylogenetic clustering of HIV from patients with recent infection. PLoS Comput Biol 2012; 8: e1002552.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Audelin AM, Cowan SA, Obel N. et al. Phylogenetics of the Danish HIV epidemic: the role of very late presenters in sustaining the epidemic. J Acquir Immune Defic Syndr 2013; 62: 102–8. [DOI] [PubMed] [Google Scholar]
- 22. Fisher M, Pao D, Brown AE. et al. Determinants of HIV-1 transmission in men who have sex with men: a combined clinical, epidemiological and phylogenetic approach. AIDS 2010; 24: 1739–47. [DOI] [PubMed] [Google Scholar]
- 23. Frentz D, Wensing AM, Albert J. et al. Limited cross-border infections in patients newly diagnosed with HIV in Europe. Retrovirology 2013; 10: 36.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grabowski MK, Lessler J, Redd AD. et al. The role of viral introductions in sustaining community-based HIV epidemics in rural Uganda: evidence from spatial clustering, phylogenetics, and egocentric transmission models. PLoS Med 2014; 11: e1001610.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hue S, Pillay D, Clewley JP. et al. Genetic analysis reveals the complex structure of HIV-1 transmission within defined risk groups. Proc Natl Acad Sci USA 2005; 102: 4425–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mehta SR, Wertheim JO, Brouwer KC. et al. HIV transmission networks in the San Diego–Tijuana border region. EBioMedicine 2015; 2: 1456–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.HyDRA Web. Public Health Agency of Canada. https://hydra.canada.ca.
- 28. Enns E, Liang B, Ji H. et al. HyDRA – a novel bioinformatics tool for next generation sequencing-based HIV drug resistance data analysis. The Canadian Association for HIV Research 25th Annual Canadian Conference on HIV/AIDS Research, Winnipeg, MB, Canada, 2016. Abstract CSP12.01.
- 29. Ji H, Enns E, Brumme CJ. et al. Bioinformatic data processing pipelines in support of next-generation sequencing-based HIV drug resistance testing: the Winnipeg Consensus. J Int AIDS Soc 2018; 21: e25193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stanford HIV Drug Resistance Database. HIVdb Program. Stanford University. http://sierra2.stanford.edu/sierra/servlet/JSierra.
- 31. Liu TF, Shafer RW.. Web resources for HIV type 1 genotypic-resistance test interpretation. Clin Infect Dis 2006; 42: 1608–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.WHO. Surveillance of HIV Drug Resistance in Adults Initiating Antiretroviral Therapy (Pre-treatment HIV Drug Resistance). Concept Note. 2014. http://www.who.int/hiv/pub/drugresistance/pretreatment_drugresistance/en/.
- 33. Bennett DE, Camacho RJ, Otelea D. et al. Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PLoS One 2009; 4: e4724.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kosakovsky Pond SL, Weaver S, Leigh Brown AJ. et al. HIV-TRACE (Transmission Cluster Engine): a tool for large scale molecular epidemiology of HIV-1 and other rapidly evolving pathogens. Mol Biol Evol 2018; 35: 1812–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ragonnet-Cronin M, Hu YW, Morris SR. et al. HIV transmission networks among transgender women in Los Angeles County, CA, USA: a phylogenetic analysis of surveillance data. Lancet HIV 2019; 6: e164–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cuypers L, Vrancken B, Fabeni L. et al. Implications of hepatitis C virus subtype 1a migration patterns for virus genetic sequencing policies in Italy. BMC Evol Biol 2017; 17: 70.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Price MN, Dehal PS, Arkin AP.. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One 2010; 5: e9490.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Guindon S, Dufayard JF, Lefort V. et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 2010; 59: 307–21. [DOI] [PubMed] [Google Scholar]
- 39. Perez AB, Vrancken B, Chueca N. et al. Increasing importance of European lineages in seeding the hepatitis C virus subtype 1a epidemic in Spain. Euro Surveill 2019; 24: pii=1800227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lemey P, Rambaut A, Drummond AJ. et al. Bayesian phylogeography finds its roots. PLoS Comput Biol 2009; 5: e1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Suchard MA, Lemey P, Baele G. et al. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol 2018; 4: vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Edwards CJ, Suchard MA, Lemey P. et al. Ancient hybridization and an Irish origin for the modern polar bear matriline. Curr Biol 2011; 21: 1251–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bielejec F, Baele G, Vrancken B. et al. SpreaD3: interactive visualization of spatiotemporal history and trait evolutionary processes. Mol Biol Evol 2016; 33: 2167–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kass RE, Raftery AE.. Bayes factors. J Am Stat Assoc 1995; 90: 773–95. [Google Scholar]
- 45. Minin VN, Bloomquist EW, Suchard MA.. Smooth skyride through a rough skyline: Bayesian coalescent-based inference of population dynamics. Mol Biol Evol 2008; 25: 1459–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Minin VN, Suchard MA.. Counting labeled transitions in continuous-time Markov models of evolution. J Math Biol 2008; 56: 391–412. [DOI] [PubMed] [Google Scholar]
- 47. Faria NR, Rambaut A, Suchard MA. et al. HIV epidemiology. The early spread and epidemic ignition of HIV-1 in human populations. Science 2014; 346: 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rambaut A, Drummond AJ, Xie D. et al. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst Biol 2018; 67: 901–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gupta RK, Gregson J, Parkin N. et al. HIV-1 drug resistance before initiation or re-initiation of first-line antiretroviral therapy in low-income and middle-income countries: a systematic review and meta-regression analysis. Lancet Infect Dis 2018; 18: 346–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.CENSIDA/Secretaría de Salud. Guía de Manejo Antirretroviral de las Personas con VIH. Décima Edición. Gobierno de México. 2019. https://www.gob.mx/censida/documentos/guia-de-manejo-antirretroviral-de-las-personas-con-vih.
- 51. Lee ER, Enns E, Parkin N. et al. Comparison of next-generation sequencing analysis pipelines for HIV-1 drug resistance. Conference on Retroviruses and Opportunistic Infections, Seattle, WA, USA, 2019. Abstract 542.
- 52. Wertheim JO, Oster AM, Johnson JA. et al. Transmission fitness of drug-resistant HIV revealed in a surveillance system transmission network. Virus Evol 2017; 3: vex008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Abram ME, Hluhanich RM, Goodman DD. et al. Impact of primary elvitegravir resistance-associated mutations in HIV-1 integrase on drug susceptibility and viral replication fitness. Antimicrob Agents Chemother 2013; 57: 2654–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hu Z, Kuritzkes DR.. Altered viral fitness and drug susceptibility in HIV-1 carrying mutations that confer resistance to nonnucleoside reverse transcriptase and integrase strand transfer inhibitors. J Virol 2014; 88: 9268–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Metzner KJ, Scherrer AU, Preiswerk B. et al. Origin of minority drug-resistant HIV-1 variants in primary HIV-1 infection. J Infect Dis 2013; 208: 1102–12. [DOI] [PubMed] [Google Scholar]
- 56. Chaillon A, Nakazawa M, Wertheim JO. et al. No substantial evidence for sexual transmission of minority HIV drug resistance mutations in men who have sex with men. J Virol 2017; 91: e00769-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gianella S, Delport W, Pacold ME. et al. Detection of minority resistance during early HIV-1 infection: natural variation and spurious detection rather than transmission and evolution of multiple viral variants. J Virol 2011; 85: 8359–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li JZ, Paredes R, Ribaudo HJ. et al. Impact of minority nonnucleoside reverse transcriptase inhibitor resistance mutations on resistance genotype after virologic failure. J Infect Dis 2012; 207: 893–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Li JZ, Paredes R, Ribaudo HJ. et al. Relationship between minority nonnucleoside reverse transcriptase inhibitor resistance mutations, adherence, and the risk of virologic failure. AIDS 2012; 26: 185–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Paredes R, Lalama CM, Ribaudo HJ. et al. Pre-existing minority drug-resistant HIV-1 variants, adherence, and risk of antiretroviral treatment failure. J Infect Dis 2010; 201: 662–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.INEGI. Encuesta Intercensal Mexico 2015. https://www.inegi.org.mx/programas/intercensal/2015/default.html#Tabulados.
- 62. Ragonnet-Cronin M, Hodcroft EB, Wertheim JO.. Understanding disclosed and cryptic HIV transmission risk via genetic analysis: what are we missing and when does it matter? Curr Opin HIV AIDS 2019; 14: 205–12. [DOI] [PubMed] [Google Scholar]
- 63. Kusejko K, Kadelka C, Marzel A. et al. Inferring the age difference in HIV transmission pairs by applying phylogenetic methods on the HIV transmission network of the Swiss HIV Cohort Study. Virus Evol 2018; 4: vey024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.