Figure 5. Identification of ADRB3 as a Target Gene of p53 R178C Using ChIP-seq Analysis.
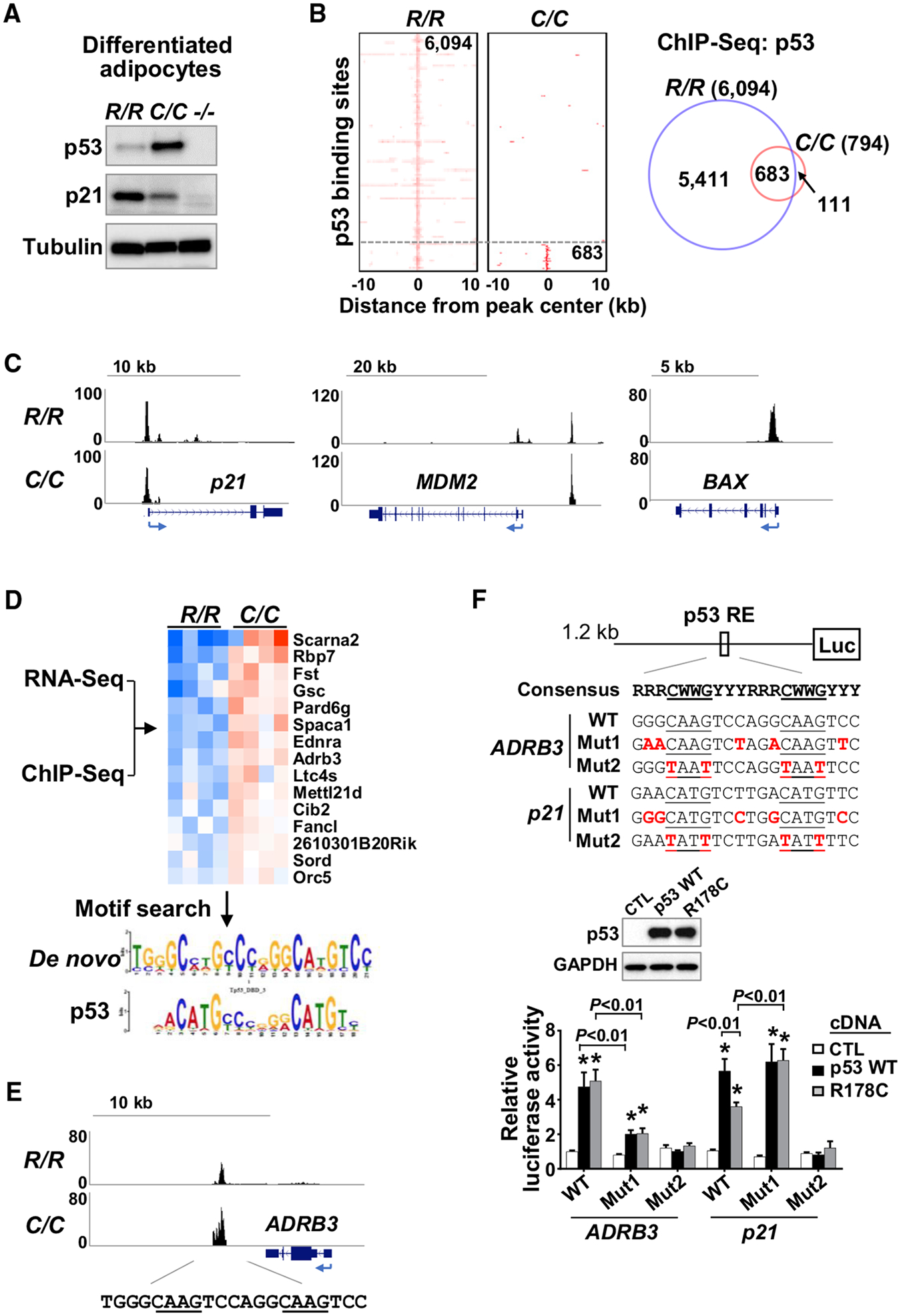
(A) Stromal vascular fractions (SVFs) isolated from iWAT of the indicated p53 genotype mice, differentiated into adipocytes and analyzed by immunoblotting.
(B) Heatmaps and Venn diagram of high-confidence (FDR < 1E 10) p53 binding sites from ChIP-seq analysis of R/R and C/C differentiated adipocytes described in (A).
(C) Genome browser view of p53 binding peaks on known wild-type p53 target genes.
(D) RNA-seq and ChIP-seq data were integrated to identify genes that are upregulated and bound by p53 R178C. p53-bound genomic sequences were subjected to a transcription factor motif search, which yielded a de novo motif (E value = 2E–44) matching the p53 binding motif (E value = 3E–5).
(E) ChIP-seq profile of the ADRB3 gene region with sequences containing the consensus p53 RE.
(F) Effect of GC-rich sequences in the p53 RE of ADRB3 on transactivation by p53, assessed by luciferase reporter assay. The GC-rich flanking sequences were mutated from GC to AT (Mut 1), and the p53 RE core sequence was also mutated as an additional negative control (Mut 2). The luciferase reporter constructs were co-transfected with wild-type p53, p53 R178C, or a control pcDNA empty vector (CTL) into p53−/− 3T3L1 cells. p53 expression levels were assessed by immunoblotting (center panel), and luciferase activity levels of the respective constructs were measured (n = 4).
p53 R178 genotypes: wild-type (R/R), homozygous mutant (C/C), and null (−/−). Statistical difference by two-tailed unpaired t test in comparison with the vector control. Values are mean ± SEM. *p < 0.05. See also Table S1.
