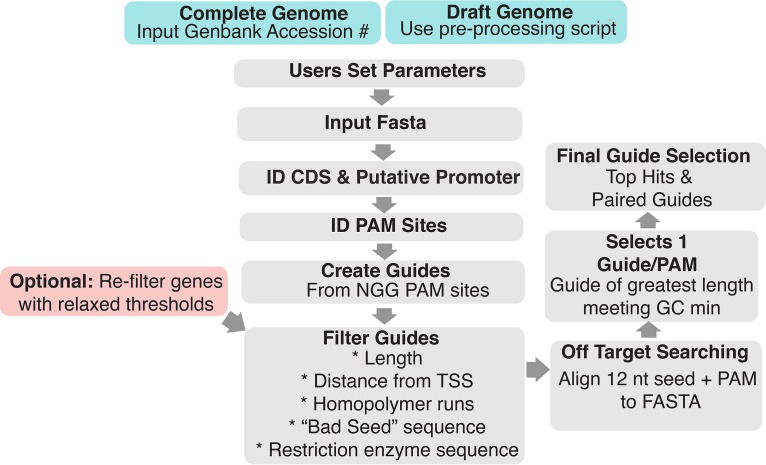
FIG 1.
GuideFinder workflow. Users set parameters and input FASTA files. Coding sequence and promoter coordinates are identified and used to obtain sequences. PAM sites are identified, and guides are created and filtered. Off-target searching is conducted using BLAST. The final guide selection step creates a top hits list and a paired guides list. Genes without guides are identified and re-run with relaxed parameters. CDS, coding DNA sequence; PAM, protospacer-adjacent motif; TSS, transcription start site; nt, nucleotide.