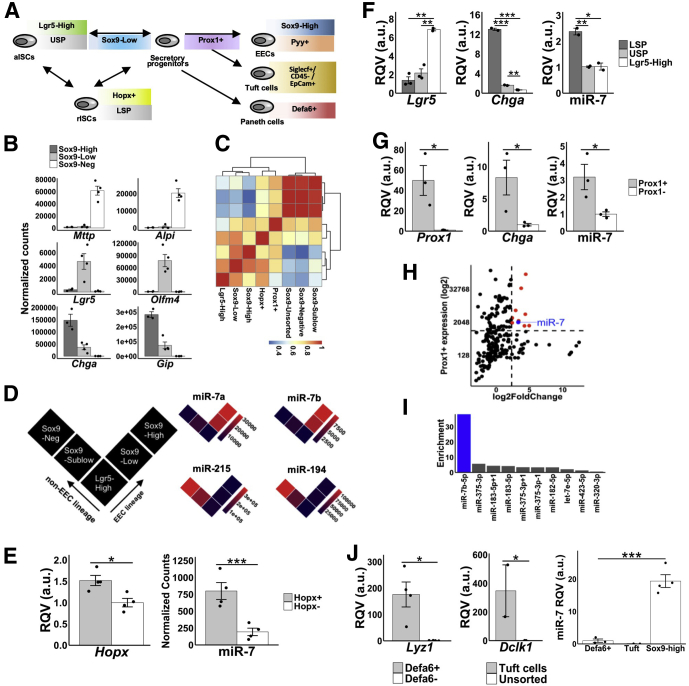
Figure 1.
MicroRNA-7 is highly enriched in the enteroendocrine (EEC) lineage trajectory. (A) Schematic diagram of different sorted cell populations representing specific cell lineages in the small intestine. (B) Level of expression (RNA-seq) of specific marker genes in each of the Sox9-Low (n = 4), Sox9-High (n = 3), and Sox9-Negative (n = 4) populations of cells. (C) Hierarchical clustering analysis based on the expression profiles of the top 50 most variable miRNAs across the different sorted cell populations shown in the heat map (Sox9-Low, n = 3; Sox9-High, n = 3; Sox9-Negative, n = 3; Sox9-Sublow, n = 3; Sox9-Unsorted, n = 2; Lgr5-High, n = 2; Hopx+, n = 4; Prox1+, n = 3). (D) MiR-7a/b expression in the EEC lineage vs non-EEC absorptive lineage. Similar data for miR-194 and miR-215 provided for sake of comparison. (E) RT-qPCR data showing enrichment of Hopx and miR-7 in Hopx+ cells (n = 4) relative to Hopx– cells (n = 4). (F) RT-qPCR data showing enrichment of miR-7, Lgr5, and Chga in LSP (n = 2) relative to USP (n = 2) and Lgr5+ cells (n = 2). (G) RT-qPCR data showing enrichment of Prox1, miR-7, and Chga in Prox1+ cells (n = 3) compared with Prox1– cells (n = 3). (H) Scatter plot showing abundance (y-axis) and enrichment (x-axis) of all detected miRNAs in Prox1+ cells (n = 3) relative to Prox1– cells (n = 3). MiRNAs above expression of 1000 reads per million mapped to miRNAs and 5-fold enrichment are shown in red (n = 10). Among these, miR-7b is highlighted in blue. (I) Fold-difference in expression of the 10 miRNAs highlighted in panel F in Prox1+ cells (n = 3) relative to Lgr5+ cells (n = 2) highlights miR-7 (blue) as a robust EEC progenitor cell enriched miRNA. (J) The left panel shows RT-qPCR data showing enrichment of Lyz1 (marker of Paneth cells) in Defa6+ (n = 4) relative to Defa6– cells (n = 4). The middle panel shows RT-qPCR data showing enrichment of Dclk1 (marker of tuft cells) in Siglecf+/CD45-/EpCam+ cells (n = 2) relative to unsorted cells (n = 2). The right panel shows RT-qPCR data showing miR-7 enrichment in EECs (Sox9-High; n = 3) compared with Paneth and tuft cells. * P < .05, ** P < .01, *** P < .001 by 2-tailed Student t test. RQV, relative quantitative value.