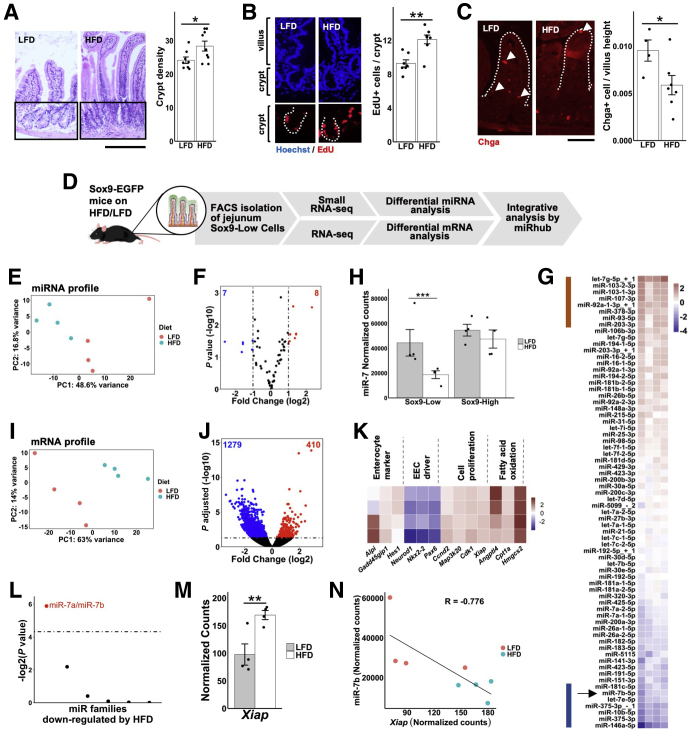
Figure 2.
MiR-7 expression is suppressed and predicted miR-7 targets are elevated in EEC progenitors under conditions of increased intestinal crypt division and reduced EEC abundance. (A) Representative hematoxylin and eosin images and crypt density quantification of mid-jejunum from HFD-fed (n = 9) and LFD-fed (n = 8) mix of Lgr5-EGFP reporter mice and naïve C57BL/6 wild-type (WT) mice. Bar = 100 μm. (B) Representative images of tissue sections stained with Hoechst (blue) and EdU (red) and quantification of EdU+ cells per crypt from HFD-fed (n = 8) and LFD-fed (n = 7) Lgr5-EGFP reporter mice and naïve C57BL/6 WT mice. (C) Representative images of tissue sections stained with Chga and quantification of Chga+ cells per villi (normalized by villi height) from HFD-fed (n = 7) and LFD-fed (n = 4) mix of Lgr5-EGFP reporter mice and C57BL/6 naïve WT mice. (D) Schematic of experimental workflow using Sox9-EGFP reporter mice. (E) Principal component analysis plot of small RNA-seq data of Sox9-Low cells from HFD-fed and LFD-fed Sox9-EGFP reporter mice. (F) Volcano plot of differentially expressed miRNAs in HFD-fed relative to LFD-fed Sox9-EGFP reporter mice (dashed lines represent fold-change > 2). (G) miRNAs that are significantly (P < .05) upregulated or downregulated in jejunal Sox9-Low sorted cells from HFD-fed relative to LFD-fed mice. Red and blue bars highlight miRNAs with >2-fold change up or down in HFD-fed relative to LFD-fed mice, respectively. (H) Expression level of miR-7b (small RNA-seq) in Sox9-Low and Sox9-High cells from HFD-fed and LFD-fed Sox9-EGFP mice. (I) Principal component analysis plot of RNA-seq data of Sox9-Low cells from HFD-fed and LFD-fed mice. (J) Volcano plot of differentially expressed genes in HFD-fed relative to LFD-fed Sox9-EGFP reporter mice (dashed line represents adjusted P < .05). (K) Heatmap showing changes in genes involved in proliferation, EEC differentiation, and enterocyte differentiation of jejunal Sox9-Low cells of HFD-fed relative to LFD-fed mice. (L) Enrichment analysis of miRNA target sites (using the Monte Carlo simulation tool miRhub) in genes upregulated in response to HFD (dashed line represents P < .05). Only miRNAs downregulated greater than 2-fold in Sox9-Low cells from HFD-fed relative to LFD-fed mice are included in the analysis. (M) Sequencing data showing upregulation of miR-7 target gene Xiap in Sox9-Low cells from HFD-fed relative to LFD-fed mice. (N) Inverse correlation between expression level of miR-7b and Xiap among the HFD-fed and LFD-fed Sox9-EGFP reporter mice. All figure panels from panel E onward correspond to data from n = 4 HFD-fed and n = 4 LFD-fed Sox9-EGFP reporter mice. * P < .05, ** P < .01 by 2-tailed Student t test.