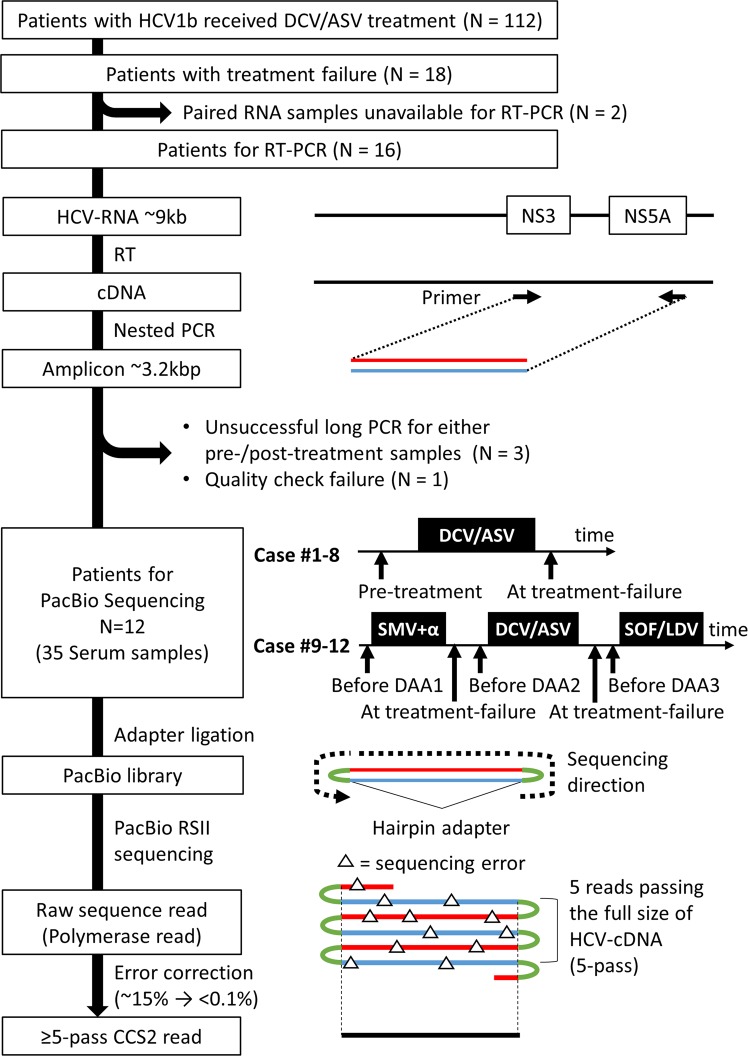
Figure 1.
Sample collection and single-molecule real-time sequencing workflow. This flow consisted of sample collection, reverse transcription polymerase chain reaction (RT-PCR), single-molecule real-time (SMRT) sequencing, and Circular Consensus Sequencing2 (CCS2). For cases #1–#8, the samples were extracted prior to treatment and after treatment failure of the daclatasvir + asunaprevir (DCV/ASV). For cases #9–#12, the samples were extracted prior to treatment with simeprevir (SMV) combined with peginterferon and ribavirin, DCV/ASV and sofosbuvir + ledipasvir (SOF/LDV), and after the treatment failure of the SMV and DCV/ASV. RT-PCR amplified the NS3-NS5A gene regions of the cDNA transcribed from HCV-RNA. For SMRT sequencing, we constructed the library by binding the adapters to the cDNA by ligation. The library was sequenced with PacBio RS II. While the sequenced raw reads included sequencing errors, CCS2, the error correction algorithm, generated highly accurate reads by correcting the errors with the raw reads repetitively sequenced in single the molecules.