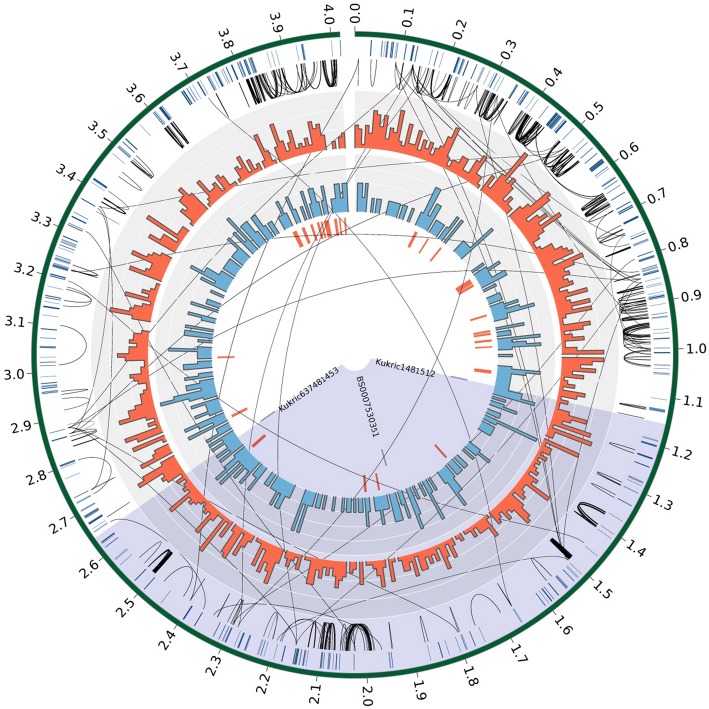
Fig. 3.
Circos plot illustrating key genomic and genic features of the ‘Synthetic W7984’ ‘super-contig’ (705 assembly scaffolds, totalling 3.47 Mb) spanning the wider Tsc2 locus, based on the ‘Chinese Spring’ region 2B: 14,040,000–30,500,000 bp. Tracks, from outside to inside: (1) the super-contig, with size indicated in Mb. (2) tick marks to illustrate start/end points between scaffolds, (3) loops to indicate sequence homology of genes based on 91-mers, (4) histogram of transposable element density, (5) histogram of gene density, (6) candidate genes, (7) tick marks indicating the named SNPs that delineate the boundaries of the most likely Tsc2 region: (a) regions kukric1481512 to kukric637481453 (full names Kukri_c148_1512 and Kukri_c63748_1453, respectively) represent a conservative Tsc2 interval, based on clear recombinations in the AM panel and encompassing the peak of the MAGIC QTL in this region, (b) region BS0007530351 (full name BS00075303_51, originating from the same RefSeqv1.1 gene model as BS00070050_51) to kukric637481453, which spans the peak of the MAGIC QTL and contains AM panel GWAS hits 1–6, 10 and 12