Abstract
Xpert MTB/RIF Ultra (Ultra) detects Mycobacterium tuberculosis and rifampicin resistance. Follow-on drug susceptibility testing (DST) requires additional sputum. Extract from the diamond-shaped chamber of the cartridge (dCE) of Ultra’s predecessor, Xpert MTB/RIF (Xpert), is useful for MTBDRsl-based DST but this is unexplored with Ultra. Furthermore, whether CE from non-diamond compartments is useful, the performance of FluoroType MTBDR (FT) on CE, and rpoB cross-contamination risk associated with the extraction procedure are unknown. We tested MTBDRsl, MTBDRplus, and FT on CEs from chambers from cartridges (Ultra, Xpert) tested on bacilli dilution series. MTBDRsl on Ultra dCE on TB-positive sputa (n = 40) was also evaluated and, separately, rpoB amplicon cross-contamination risk . MTBDRsl on Ultra dCE from dilutions ≥103 CFU/ml (CTmin <25, >“low semi-quantitation”) detected fluoroquinolone (FQ) and second-line injectable (SLID) susceptibility and resistance correctly (some SLIDs-indeterminate). At the same threshold (at which ~85% of Ultra-positives in our setting would be eligible), 35/35 (100%) FQ and 34/35 (97%) SLID results from Ultra dCE were concordant with sputa results. Tests on other chambers were unfeasible. No tubes open during 20 batched extractions had FT-detected rpoB cross-contamination. False-positive Ultra rpoB results was observed when dCE dilutions ≤10−3 were re-tested. MTBDRsl on Ultra dCE is concordant with isolate results. rpoB amplicon cross-contamination is unlikely. These data mitigate additional specimen collection for second-line DST and cross-contamination concerns.
Subject terms: Tuberculosis, Laboratory techniques and procedures, Translational research, Molecular medicine
Introduction
Drug-resistant tuberculosis (TB) remains a global threat1. Of 10 million estimated incidence cases reported in 2017, 588 000 were rifampicin-resistant2. Of these ~458 000 were multidrug-resistant (MDR). Despite the improved roll-out of rifampicin-resistance testing, many patients are not diagnosed appropriately or started on effective treatment, resulting in huge TB care cascade gaps3,4. For example, in South Africa, 84% of patients with drug-resistant TB have access to rifampicin-susceptibility testing, but only 47% of these are started on likely effective treatment4. Similarly, in India, only 41% of the MDR-TB burden was diagnosed in 2013 and, of these, just 32% started on treatment5. Innovative approaches are needed to ensure more patients receive comprehensive drug susceptibility testing (DST).
Previous work showed that mycobacterial genomic DNA can be recovered from the rear diamond-shaped chamber of used Xpert MTB/RIF (Xpert) cartridges after the test is complete. This diamond cartridge extract (dCE) is useful for downstream testing with the MTBDRsl line probe assay (LPA) (Hain Lifescience, Germany), the only World Health Organization (WHO)-endorsed molecular test for second-line drug resistance, and spoligotyping6, a method useful for monitoring the molecular epidemiology of TB outbreaks. This additional testing does not require extra specimen collection nor additional downstream DNA extraction, both of which can exacerbate patient loss within the diagnostic care cascade.
As Xpert is a real-time PCR that generates quantitative information, a cycle threshold value (CT <24) was identified at which downstream dCE testing using MTBDRsl was successful and fully concordant with MTBDRsl results on matching isolates7. However, Xpert dCE was not useful for first-line DST using the WHO-endorsed MTBDRplus assay, likely due to interference from large numbers of Xpert rpoB amplicons. In addition to the dCE approach, others8,9 have shown it is possible to test leftover specimen-sample reagent mix remaining after Xpert, however, remnant volume is not always present and DNA extraction and downstream clean-up might still be needed.
Xpert MTB/RIF Ultra (Ultra) recently superseded Xpert as WHO-endorsed frontline molecular test-of-choice for TB and rifampicin resistance10. Compared to Xpert, Ultra has higher sensitivity in paucibacillary samples, however, specificity is overall lower11–13. Ultra is a different assay compared to Xpert and it is not necessarily given that the extract approach would be feasible on Ultra dCE. We aimed to confirm that Ultra dCE would be useful for second-line DST. Furthermore, we asked if extract from other chambers within the cartridge other than the diamond (i.e., chambers that are likely rpoB amplicon-free), may contain DNA. We quantified this DNA using a Mycobacterium tuberculosis complex 16S rRNA real time qPCR and evaluated whether this DNA was useful for first-line DST using the FluoroType MTBDR (FT) (Hain Lifescience, Germany) assay14,15. A test such as FT could, for example, be used to check for isoniazid mono-resistance or confirm Ultra rifampicin-resistance results.
Lastly, as the cartridge extraction (CE) procedure involves aspirating fluid rich in rpoB amplicons, it may represent a source of cross-contamination. We sought to evaluate this risk, both under a prolonged exposure scenario (where collection tubes were purposely exposed during extended batch extractions) and an absolute worst-case scenario (directly adding dCE to a sample later tested by Ultra). Showing that the extracted cartridge approach in Ultra is compatible with MTBDRsl and represents minimal rpoB amplicon cross-contamination risk would increase the likelihood of implementation, especially as Xpert is in the process of being phased out in lieu of Ultra. In turn, this could reduce both sputum collection requirements for complete DST and time-to-effective-treatment initiation.
Methods
Ethics statement
Methods and protocols were carried out in accordance with relevant guidelines and regulations. The study was approved by the Health Research Ethics Committee of Stellenbosch University (N09/11/296) and the City of Cape Town (10570). Permission was granted to use anonymised residual specimens collected during routine diagnostic practice and thus patient informed consent was waived.
Ultra and Xpert on dilution series of Mycobacterium tuberculosis strains
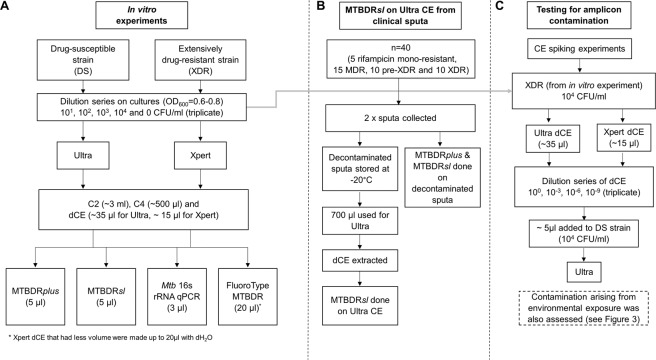
Culturing of genotypically-confirmed drug-susceptible (DS-TB) and extensively-drug resistant (XDR) M. tuberculosis isolates were done in a Biosafety Level (BSL) 3 laboratory to an OD600 of 0.6–0.8 (Fig. 1A). A triplicate tenfold dilution series from three separate cultures [100–104 colony forming units (CFU)/ml] was prepared in phosphate buffer (33 mM Na2HPO4, 33 mM KH2PO4; pH 6.8) with 0.025% Tween80 (Sigma-Aldrich, United States). Colony counts were done on 7H11 Middlebrook agar (BD Biosciences, United States). A total of 52 dilutions [four dilutions, 101–104 CFU/ml in triplicate for both strains plus a negative control for each strain; (4 × 3 × 2 + 2) × 2] were made up to 1 ml and tested by Ultra (n = 26) or Xpert (n = 26) per the manufacturer’s instructions16,17. Used positive cartridges were stored prior to extraction at 4 °C for ≤3 days. Crude DNA (heat inactivated for 2 hours at 100 °C) from the same strains served as positive controls for downstream tests (16S rRNA gene qPCR, MTBDRplus, MTBDRsl, FT).
Figure 1.
Study flow diagrams for the (A) in vitro experiment, (B) MTBDRsl on Ultra CE from clinical sputa experiment, and the (C) evaluation of rpoB amplicon cross-contamination risk experiment.
Ultra on sputum from TB patients
Forty used positive Ultra cartridges done on NALC-NaOH decontaminated sputa from pre-treatment TB patients with known drug resistance [5 rifampicin-mono-resistant, 15 MDR, 10 pre-XDR (resistance to rifampicin, isoniazid and either a fluoroquinolones or a second-line injectable), 10 XDR] were collected from November 2015 to September 2017 and dCEs were extracted as described previously6 (Fig. 1B). To confirm MTBDRsl results from dCEs, MTBDRsl was done per the manufacturer’s instructions directly on corresponding decontaminated sputa18,19. Ultra cartridges were processed in a manner blinded to MTBDRsl results.
Recovery of mycobacterial genomic DNA from used Ultra and Xpert cartridges
Preparation of work space
BSL2 hood surfaces were sterilised [1% NaOCl (bleach), 70% EtOH, 5 min UV irradiation] before and after each batched extraction. Each cartridge was wiped with 1% bleach and 70% EtOH before and after each extraction.
Description of cartridge design
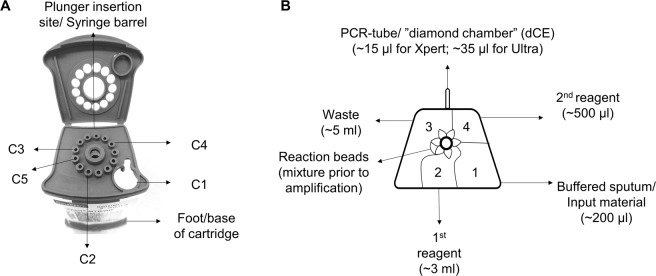
To investigate the feasibility of testing extract from Ultra and Xpert cartridge chambers, an understanding of their design and inner processes is required. As described previously, each cartridge has a similar design consisting of a foot, valve, body, reaction tube and lid20,21. The five internal chambers hold buffers and lyophilised PCR reagents used for sputum homogenisation, washing away debris, and DNA extraction, purification, and amplification22. The Xpert and Ultra procedures, including the processes inside the cartridges and the contents of each chamber are described in the supplement. After assay conclusion, the volumes typically remaining in each chamber are ~500 µl for Chamber 1 (C1), ~3 ml for Chamber 2 (C2), ~5 ml for Chamber 3 (C3) and ~500 µl for Chamber 4 (C4) [Chamber 5 (C5) had no volume remaining after test completion].
Diamond chamber extract
dCEs were extracted from all positive cartridges by puncturing the rear chamber with a sterile 29 G × 1/2′′ 1 ml insulin syringe (Avacare, South Africa) (Fig. 2A,B) as described previously6. The full volume was extracted (~15 µl for Xpert; ~35 µl for Ultra). CEs were stored in microcentrifuge tubes at −20 °C prior to analysis.
Figure 2.
(A) Entry points through the lid of the cartridge for access to different cartridge chambers. (B) Top-down cross-section of the inside of the cartridge corresponding to the access points.
Other chambers
Five cartridge chambers (C1, C2, C3, C4, C5) were accessed by inserting a 22 G spinal needle (Becton Dickinson, United States) fixed a 5 ml syringe (Fig. 2A; a pipette may also be used for C1) and the entire volume withdrawn (Fig. 2A,B). C5 had no remaining volume left after Xpert or Ultra test completion. No DNA extraction or purification steps were done for downstream assays.
16s rRNA gene quantitative PCR (qPCR) on cartridge extract
CEs from C1–4 and dCE from Ultra and Xpert done on the serial dilutions were tested (heat extracted crude DNA from matching isolates was used as positive control). For each qPCR, 5 µl iTaq Universal SYBR Green Supermix (Bio-Rad), 0.3 µl (300 nM) of M. tuberculosis specific forward (V4 515F) primers, 0.3 µl (300 nM) of M. tuberculosis specific reverse (V4 806R) primers (Table S1) and 1.4 µl nuclease-free water was used23. 3 µl CE was added and amplification occurred using a Bio-Rad CFX-96. The threshold used to determine if a reaction was excluded from subsequent analyses was defined as a Cq value greater than the average of the triplicate negative controls for that run. Chambers with a Cq less than that average value were considered positive for M. tuberculosis complex (MTBC) DNA and used for MTBDRplus, MTBDRsl and FT.
MTBDRplus and MTBDRsl line probe assays on cartridge extract
Diamond chamber extract
MTBDRplus and MTBDRsl (both version 2.0) were performed on dCEs from Ultra and Xpert done on the in vitro dilution series. For Ultra done on sputa from patients, only MTBDRsl was done. 5 µl dCE was used for MTBDRplus and MTBDRsl each. MTBDRplus and MTBDRsl results were reported as described24: either actionable [TUB-band positive and determinate (gene-specific locus bands present)] or non-actionable [TUB-band negative or TUB-band positive but indeterminate (gene-specific locus band absent)]. Susceptibility calls were made for all actionable results. Banding patterns were read by two experienced independent readers blinded to each other’s calls, the Ultra and Xpert results, and, for the dilution series experiement, the strain antibiograms (if there was a discrepancy between readers, a third experienced reader reviewed results and did the final classification).
Other chambers
MTBDRplus and MTBDRsl were done on C2 and C4 CEs from both Ultra and Xpert done on the dilution series. C1, C3, and C5 were not tested with LPAs as their CEs were 16S rRNA qPCR-negative or there was no volume remaining to test after the Ultra or Xpert test had completed (C5).
FluoroType MTBDR on cartridge extract
Diamond chamber
dCEs from Ultra and Xpert cartridges done on the in vitro dilution series were tested by FT using the manufacturer’s instructions25. A total of 26 tubes for each test (Ultra, Xpert) were tested [four dilutions from 101–104 CFU/ml in triplicate for both strains plus a negative control for each strain, (4 × 3 × 2 + 2)]. As Xpert dCE had a volume of ~15 µl, after MTBDRplus (5 µl), MTBDRsl (5 µl), and the 16S rRNA qPCR (3 µl) were all done on the same Xpert dCE, the remaining volumes (5–14 µl) were made up to 20 µl with dH2O for FT (the recommended input volume)25. All Ultra dCEs (~35 µl originally) had 20 µl remaining and the full 20 µl was used for FT. FT results were classified in a manner similar to that for the line probe assays: actionable (MTBC detected; rifampicin and isoniazid susceptible or resistant) or non-actionable (no MTBC detected, MTBC indeterminate or MTBC detected but rifampicin or isoniazid indeterminate).
Other chambers
FT was done on C2 and C4 (as for LPAs) from both Ultra and Xpert cartridges used for the dilution series.
Evaluation of rpoB amplicon cross-contamination risk
Amplicon escape during batched cartridge extractions
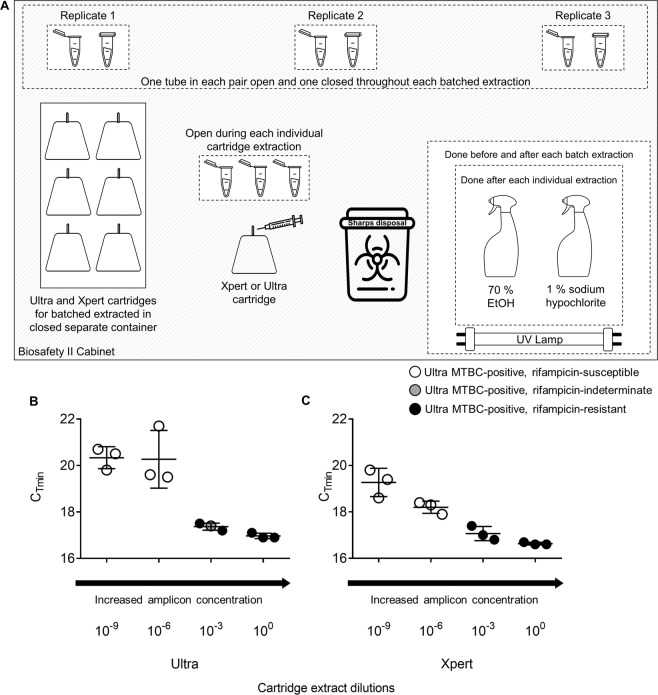
During all Ultra and Xpert diamond chamber extractions, 1.5 ml microcentrifuge tubes containing 100 µl sterile dH2O were positioned in the same BSL2 cabinet (Fig. 3A). Three tubes remained open throughout all extractions for each batch extraction and three remained closed (negative controls). Tubes were stored at −20 °C for later FT testing. A total of 20 batches of cartridges were extracted [n = 120 tubes in total from the 20 batches, n = 60 open tubes and n = 60 closed tubes including triplicates], with a median (IQR) number of cartridges per batch of 17.5 (10.5–27.5). There were also three tubes open for each individual cartridge extraction but these were not tested further based on results of the open tubes during batched extraction, which revealed no cross-contamination. Furthermore, extractions procedures were done by a total of five different users to reflect user variability.
Figure 3.
Evaluation of rpoB cross contamination risk experimental set-up and results. (A) Configuration of the environmental exposure experiment within a Biosafety level 2 cabinet. Three microcentrifuge tubes were open throughout each batched extraction procedure and three remained closed [median (IQR) extractions per batch 17.5 (10.5–27.5)]. No exposed tubes were FT rpoB-positive. In parallel to evaluate if, in an absolute worst case scenario, rpoB cross-contamination was probable, dCE from a (B) Ultra or (C) Xpert done on a drug-resistant strain was added to a drug-susceptible strain and the resultant mixture tested by Ultra. When samples of DS-TB contained CE at higher concentrations (undiluted, 10−3), false-resistant (solid black circles) or indeterminate rifampicin resistance (grey circles) are seen. All samples containing CE dilutions beyond 10−6 showed true rifampicin susceptibility (white circles). Error bars represent CTmin values for each dilution. Some images were obtained from the Noun Project: microcentrifuge tube (without changes), Anthony Ledoux, https://thenounproject.com/term/eppendorf/1699532/; spray bottle (without changes), John Winowiecki, https://thenounproject.com/search/?q=spray%20bottle&i=2236898; sharps container, Juicy Fish (with changes), https://thenounproject.com/term/hospital-waste-bin/2450390/; needle (without changes), Creative Mania; https://thenounproject.com/search/?q=injection&creator=2251916&i=2409865.
Spiking of amplicons
The same XDR-TB strain with known Xpert and Ultra rpoB resistance profiles was used in the dilution series (Fig. 1C). Ultra and Xpert were each done on 1 ml of a 104 CFU/ml concentration (in triplicate). dCEs were extracted and used for a dilution series (100, 10−3, 10−6, and 10−9; each 1 ml final volume). For all dilutions, 5 µl was added to 700 µl of the DS-TB strain (104 CFU/ml) and tested with Ultra [700 µl was used as, when combined with the recommended two-fold sample reagent volume, the 2 ml input volume is reached with minimal sample unused (~100 µl)].
Results
Mycobacterium tuberculosis complex genomic DNA detection in different chambers from cartridges done on dilution series
Though qPCR-positive results were obtained from C2, C4 and the dCE (Fig. S1), these results were highly variable even at high concentrations of bacilli (at least 104 CFU/ml), suggesting interference. As C2, C4 and dCE gave positive qPCR results on cartridges done on some dilutions, and C1 and C3 gave none, we only explored the utility of the former for downstream testing using MTBDRplus, MTBDRsl, and FT.
MTBDRplus and MTBDRsl on extract from cartridges done on dilution series
TB detection
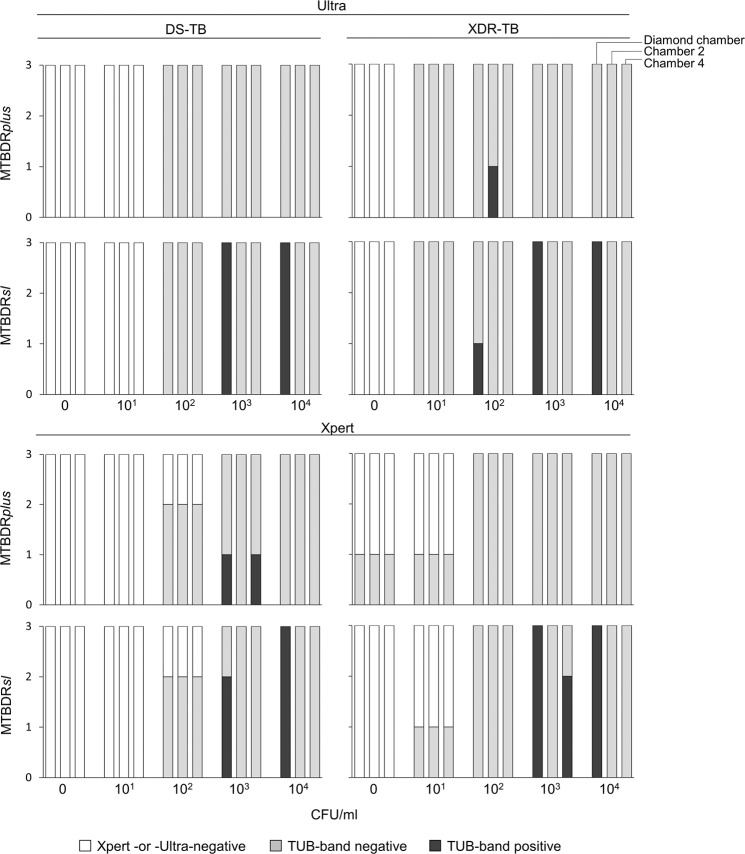
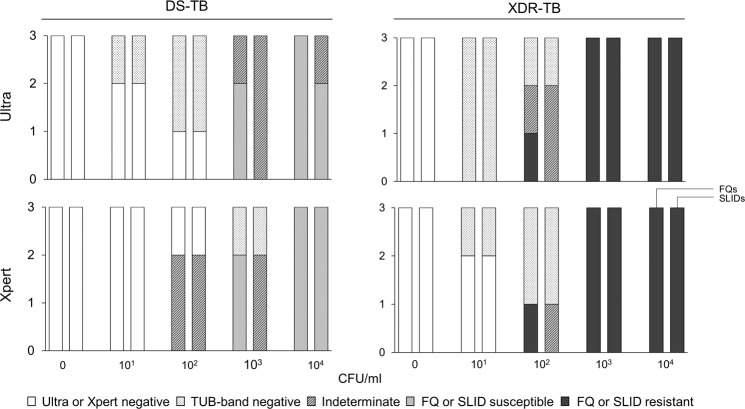
More Ultras were MTBC-positive at lower CFU titres than Xpert [e.g., 4/6 (67%) of the 101 CFU/ml aliquots vs. 1/6 (17%) for Xpert at the same concentration for both strains] (Fig. 4). MTBDRplus had high rates of non-actionable results across all dilutions irrespective of the cartridge chamber extract originated from (diamond, C2, C4) or initial test (Ultra, Xpert) (Fig. 4). MTBDRsl had actionable results for all Ultra dCEs ≥103 CFU/ml and, for Xpert, all but one dCE ≥103 CFU/ml (one Xpert replicate at 103 CFU/ml was MTBDRsl-non-actionable). MTBDRsl on C2 and C4 had non-actionable results across all dilutions (Ultra and Xpert).
Figure 4.
MTBDRplus and MTBDRsl on cartridge extract results for TB detection. dCE (left-most column), C2 (middle column) and C4 (right-most column) from M. tuberculosis-positive cartridges on dilution series (DS-TB and XDR-TB strains) are shown. MTBDRplus had mostly non-actionable results (not positive or negative). MTBDRsl had actionable results on all Ultra- and Xpert-positive dCE at >103. Though some actionable line probe assay results for non-diamond chambers were observed, these were inconsistent and had low reproducibility.
Resistance detection
MTBDRsl correctly identified FQ and SLID resistance on Ultra dCE done on all XDR strain aliquots ≥103 CFU/ml (Fig. 5). On the DS-TB strain, MTBDRsl identified FQ susceptibility in all three 104 CFU/ml replicates and in 2/3 (67%) replicates for SLIDs (one indeterminate). At 103 CFU/ml for the DS-TB strain, 2/3 (67%) were correctly identified as FQ susceptible (one indeterminate) and all were SLID-indeterminate. The CTmin threshold at which all MTBDRsl results was feasible on Ultra CE was <25, which was used for further experiments. Similar results were obtained for MTBDRsl on Xpert dCE.
Figure 5.
MTBDRsl drug susceptibility results on dCEs from Ultra and Xpert on dilution series. All results ≥103 CFU/ml for the XDR-TB strain had resistance results concordant with the isolate. Some SLIDs indeterminate results were seen for the DS-TB >103 at the same concentrations but MTBDRsl results were otherwise concordant with those on the isolate.
MTBDRsl on extract from cartridges done on clinical specimens
TB detection
As MTBDRplus was not feasible in the in vitro assessment, it was not done on CE from Ultras done on clinical sputa. MTBDRsl on dCE from Ultra done on clinical sputa had 37/40 (93%) actionable results (the rest were non-actionable). Non-actionable results corresponded to “trace” or “very low” semi-quantitative categories.
Resistance detection
Of the actionable results, 35/37 (95%) fell within the defined threshold (CTmin <25) and of these all FQ results were concordant with MTBDRsl on sputum and all but one SLID result were concordant (false-susceptible). Though this percentage is slightly higher than the number of patients with CTmin <25 in our setting, which was determined to be 86% (based on an evaluation of Ultra done in sympotmatic patients in primary care26), which further show that this approach would benefit the majority of patients in our setting. Of the 2/37 (5%) results that were actionable but fell above the defined threshold, one was concordant with MTBDRsl on sputa and one was indeterminate for FQs and discordant for SLIDs (false-resistant).
Receiver operator curve for determining actionable results
An Ultra rpoB CTmin threshold of <25.4 was defined for dCEs done on clinical sputa with sensitivities of 97% (95% CI 87–100) and specificities of 100% (55–100) (Fig. 6).
Figure 6.
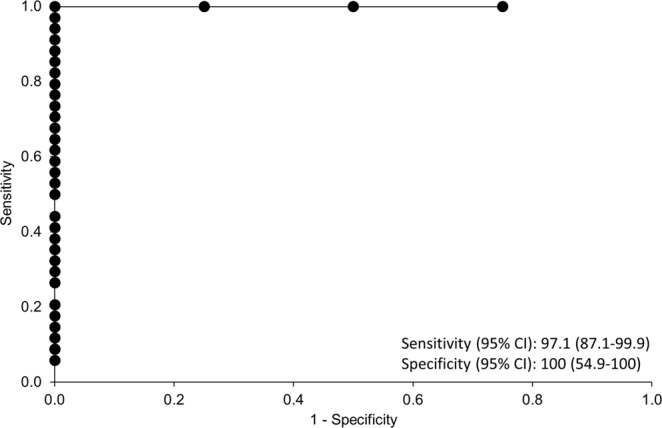
Receiver operation area under the curve of actionable vs. non-actionable results of MTBDRsl on Ultra diamond cartridge extract done on DR-TB clinical sputa to determine a CTmin threshold at which this approach is not feasible. MTBDRsl yields actionable results on cartridge extract from Ultra at a CTmin threshold of <25.4 with a sensitivity of 97% (87.1–99.9; 95% CI) and specificity of 100% (54.9–100; 95% CI) respectively.
FluoroType MTBDR on extract from cartridges done on dilution series
Diamond chamber
FT had similar results to MTBDRplus on CEs. For example, 3/24 (12%) Ultra dCEs were MTBC-positive (the others negative) for both strains (Fig. S2). In the three Ultra dCEs with a TB-positive FT result, all had indeterminate susceptibility results for at least one drug. A total of 18/24 (75%) Xpert dCEs were FT MTBC-positive, however, of these 13/24 (54) were indeterminate for at least one drug.
Chamber 2
FT on Ultra C2 had MTBC positivity rates of 10/12 (83%) and 11/12 (92%) for DS-TB and XDR-TB, respectively. On Xpert C2, FT TB positivity rates were 5/12 (42%) and 7/12 (58%) for DS-TB and XDR-TB, respectively. In MTBC-positive extracts (Ultra and Xpert), most resistance calls were indeterminate or discordant with the paired isolate.
Chamber 4
FT done on C4 from Ultra had 8/12 (67%) and 9/12 (75%) TB positivity rates for DS-TB and XDR-TB strains respectively, and 3/12 (25%) and 1/12 (8%) on for C4 from Xpert respectively. As for C2, resistance calls were mainly indeterminate or discordant with paired isolate.
rpoB amplicon cross-contamination risk evaluation
Exposure of open tubes during batched extractions
All sixty tubes exposed were FT MTBC-negative and had no rpoB amplification.
Amplicon spiking for absolute worst-case cross-contamination scenario
Of the Ultra dCEs done on XDR-TB and spiked into DS-TB for re-testing with Ultra, evidence of cross-contamination was seen when dCEs were diluted less than 10−6 before addition to the DS-TB strain [3/3 (100%) of 100 dilutions and 2/3 (67%) of the 10−3 dilutions showed false-resistance (1/3 of the 10−3 was resistance indeterminate)] (Fig. 3B). Similar results were obtained for Xpert dCE (Fig. 3C).
Discussion
We have validated MTBDRsl on CEs from used Ultra cartridges for genotypic second-line DST. We show: (1) MTBDRsl on Ultra dCE when CTmin <25 enabled DST concordant with sputum results, (2) risk of rpoB extract cross-contamination is unlikely if standard aseptic protocols are followed, (3) neither 16S rRNA qPCR, MTBDRplus, MTBDRsl nor FT are feasible on other cartridge chambers, nor was MTBDRplus or FT on Ultra and Xpert dCEs. These data support the use of Ultra extract for second-line genotypic DST.
We defined a threshold at which MTBDRsl is likely to work on Ultra dCE from the vast majority of Ultra-positive patients, thereby avoiding time and resources wasted on dCE unlikely to give a valid result. We are mindful that there were some indeterminate SLID results (in line with previous reports of higher MTBDRsl indeterminate result rates for SLIDs vs. FQs)27–29. However, all dCE SLID-indeterminate results from the dilution series were from the DS-TB strain and there were no indeterminate SLID results on XDR-TB dCEs. On clinical sputum (and falling within our threshold), one MTBDRsl SLID susceptibility result was discordant with sputum (one false-negative). We thus suggest that MTBDRsl Ultra dCE results are interpreted in the same manner as recommended by the WHO for MTBDRsl on clinical specimens30. If, for example, MTBDRsl on dCE is non-actionable or susceptible, MTBDRsl on sputum or isolates should be done. If there is still no evidence of resistance in a high burden setting, phenotypic DST should still be done given the suboptimal rule-out accuracy of MTBDRsl19,30.
The possibility of contamination from rpoB amplicons during extractions has not been investigated. We implemented systematic testing for possible environmental contamination. No tubes exposed for each extraction batch were rpoB-positive when tested with FT. FT was used for testing for rpoB amplicons as it is more sensitive than MTBDRplus14,15.
We further tested a worst-case contamination scenario with dCEs from both Ultra and Xpert cartridges done on a XDR-TB strain, diluting these dCEs, and adding them to a DS-TB strain which was subsequently tested by Ultra. The undiluted and most concentrated dCE dilutions (100, 10−3) showed false rifampicin-resistance indicating that, although the GeneXpert platform does have proven ability to remove large numbers of amplicons31, it was not able to remove all amplicons during the pre-amplification wash steps, however, amplicons diluted beyond 10−3 were successfully removed to the point of not being detected22,32,33. These results, together with those from the environmental samplings during extractions, shows that when standard aseptic techniques are used, amplicon cross-contamination is highly unlikely except in the artificial worst case scenarios. Finally, it should be noted that, in line with good practice in any molecular biology laboratory providing results for patient management, dCEs should not be collected in the same room where rpoB-based tests are done, and that the risk of cross-contamination from the dCE approach is only pertinent to tests for rifampicin resistance.
We suggest that diagnosticians considering implementing this approach use the cartridge itself as a transport vessel (upright and in sealed containers) to a central laboratory where dCE can be extracted appropriately (the diamond is a sealed chamber and should remain safe during transport). Most peripheral laboratories will be unable to do the dCE procedure safely and downstream molecular DST like MTBDRsl. This cartridge transport can interface with existing specimen referral networks. If dCE is planned purely for molecular epidemiology, we suggest that dCE be extracted and stored at −80 °C or alternatively the whole cartridge be stored at −20 °C until extractions can be done in a batched, centralised fashion. The long term stability of these approaches will require examination.
We further hypothesised that liquid from other cartridge chambers may avoid interference by rpoB amplicons. However, upon testing, this approach gave variable non-replicable results. This was true for qPCR, MTBDRplus, MTBDRsl and FT assays. This may also be due to very low concentrations of template in these chambers, for example C3 – which is the “wash chamber”, and/or remnant PCR inhibitors (e.g., salts from the sample reagent). In light of this, we believe that the presence of these amplicons may prevent newer approaches, such as next generation sequencing methods, from performing well on dCE without to clean up steps. This warrants further investigation. CE from the diamond chamber hence remains the best option for downstream genotypic DST.
The results of this study should be interpreted within its limitations, namely aseptic techniques done in an assay- or procedure-specific biosafety cabinet are needed to minimise amplicon cross-contamination. However, this infrastructure should already be implemented per WHO guidelines34 where LPAs are done routinely for patient care. Furthermore, per good laboratory practice, CEs should not be collected in the same room where rpoB- or IS6110/1081-based assays are done, nor should either procedure be done by the same personnel on a daily basis. Lastly, further investigation into cross-contamination risk should be done in a routine diagnostic setting. This should include multiple operators.
We also acknowledge that this method may increase risk of needle stick injury. Standard biosafety protocols should be strictly adhered to. We were recently funded to develop a device that can eject material from cartridges in a safe manner. Another limitation is MTBDRplus was not feasible on Ultra CEs and we suspect this is due to interference from both rpoB and IS6110/1081 amplicons. Thus, combined with the large volumes (and hence diluted targeted DNA) recovered from non-diamond chambers in Ultra and Xpert, MTBDRplus (and also likely FT) on extract from any Ultra cartridge chamber is in all likelihood not useful for isoniazid or confirmatory rifampicin DST. Finally, although the diamond chamber is a closed system and appears protected against desiccation, we acknowledge that some desiccation may occur over prolonged periods that this requires future systematic evaluation. However, we recommend that extract method is done on an as fresh a cartridge as possible (either at a peripheral or central laboratory), in order to reduce the delays of DR-TB diagnosis. Formal evaluation of CE stability pre-extraction may be useful.
We conclude that dCEs from Ultra at the CTmin threshold (<25), can be used for genotypic second-line DST (MTBDRsl). Ultra and MTBDRsl on dCE therefore allows for the rapid rule-in detection of XDR-TB on a single specimen.
Supplementary information
Acknowledgements
The authors would like to sincerely thank Roxanne Higgit for her assistance in the extraction process as well as Dr. Charissa Naidoo for assistance with the qPCR protocol and Dr Anzaan Dippenaar for her assistance with the FT assays. The financial assistance of the National Research Foundation (NRF) towards this research is hereby acknowledged. Opinions expressed and conclusions arrived at, are those of the author(s) and are not necessarily to be attributed to the NRF. Research reported in this publication was supported by the South African Medical Research Council. The content is solely the responsibility of the authors and does not necessarily represent the official views of the South African Medical Research Council. GT acknowledges funding from South African Medical Research Council (SAMRC Flagship Project MRC-RFA-IFSP-01-2013), the EDCTP2 program supported by the European Union (grant SF1401, OPTIMAL DIAGNOSIS), and the Faculty of Medicine and Health Sciences, Stellenbosch University. This research was supported by The Center for Innovation in Point-of-Care Technologies for HIV/AIDS at NorthwesternUniversity. The Research reported in this publication was supported by the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health under Award Number U54EB027049. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions
G.T., R.W., M.D.V. and R.V. conceived the experiments. R.V., S.M., B.D., H.T., and A.R. conducted the experiments. T.D. provided specimens and data from the NHLS. R.V. and S.M. analysed data. All authors reviewed the manuscript.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on request.
Competing interests
G.T. acknowledges in-kind donations from Hain Lifesciences. G.T. acknowledges in-kind donations from Cepheid for other studies. G.T., R.W., and M.d.V. acknowledge in-kind donations and funding from Hain Lifesciences for other studies. G.T., R.W. and M.d.V. declare no other competing interests. The other authors each declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Rouxjeane Venter and Stephanie Minnies.
Supplementary information
is available for this paper at 10.1038/s41598-020-59164-3.
References
- 1.Dheda K, et al. The epidemiology, pathogenesis, transmission, diagnosis, and management of multidrug-resistant, extensively drug-resistant, and incurable tuberculosis. The lancet Respiratory medicine. 2017;5:291–360. doi: 10.1016/S2213-2600(17)30079-6. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. Global Tuberculosis Report (2019).
- 3.Gandhi NR, et al. Multidrug-resistant and extensively drug-resistant tuberculosis: a threat to global control of tuberculosis. The Lancet. 2010;375:1830–1843. doi: 10.1016/S0140-6736(10)60410-2. [DOI] [PubMed] [Google Scholar]
- 4.Naidoo P, et al. The South African Tuberculosis Care Cascade: Estimated Losses and Methodological Challenges. The Journal of infectious diseases. 2017;216:S702–S713. doi: 10.1093/infdis/jix335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Subbaraman R, et al. The tuberculosis cascade of care in India’s public sector: a systematic review and meta-analysis. PLoS medicine. 2016;13:e1002149. doi: 10.1371/journal.pmed.1002149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Venter R, et al. Mycobacterial genomic DNA from used Xpert MTB/RIF cartridges can be utilised for accurate second-line genotypic drug susceptibility testing and spoligotyping. Scientific reports. 2017;7:14854. doi: 10.1038/s41598-017-14385-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Theron G, et al. The Use of an Automated Quantitative Polymerase Chain Reaction (Xpert MTB/RIF) to Predict the Sputum Smear Status of Tuberculosis Patients. Clinical Infectious Diseases. 2012;54:384–388. doi: 10.1093/cid/cir824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alame-Emane, A. K. et al. The use of GeneXpert remnants for drug resistance profiling and molecular epidemiology of tuberculosis in Libreville, Gabon. Journal of Clinical Microbiology, JCM. 02257–02216 (2017). [DOI] [PMC free article] [PubMed]
- 9.Mambuque, E. T. et al. Direct genotyping of Mycobacterium tuberculosis from Xpert® MTB/RIF remnants. Tuberculosis (2018). [DOI] [PubMed]
- 10.World Health Organization. WHO meeting report of a technical expert consultation: Non-inferiority analysis of Xpert MTB/RIF Ultra compared to Xpert MTB/RIF. (2017).
- 11.Chakravorty S, et al. The new Xpert MTB/RIF Ultra: improving detection of Mycobacterium tuberculosis and resistance to rifampin in an assay suitable for point-of-care testing. MBio. 2017;8:e00812–00817. doi: 10.1128/mBio.00812-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorman SE, et al. Xpert MTB/RIF Ultra for detection of Mycobacterium tuberculosis and rifampicin resistance: a prospective multicentre diagnostic accuracy study. The Lancet Infectious Diseases. 2018;18:76–84. doi: 10.1016/S1473-3099(17)30691-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mishra, H. et al. Diagnostic accuracy and predictive value of Xpert Ultra and Xpert MTB/RIF for tuberculosis diagnosis in an HIV-endemic setting with a high burden of previous tuberculosis Lancet Respiratory Medicine (2019). [DOI] [PubMed]
- 14.de Vos M, et al. Diagnostic accuracy and utility of FluoroType MTBDR, a new molecular assay for multidrug-resistant tuberculosis. Journal of clinical microbiology. 2018;56:e00531–00518. doi: 10.1128/JCM.00531-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hillemann D, Haasis C, Andres S, Behn T, Kranzer K. Validation of the FluoroType MTBDR assay for detection of rifampin and isoniazid resistance in Mycobacterium tuberculosis complex isolates. Journal of clinical microbiology. 2018;56:e00072–00018. doi: 10.1128/JCM.00072-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.World Health Organization. Xpert MTB/RIF implementation manual: technical and perational ‘how-to’: practical considerations [Internet]. Geneva: World Health Organization. (2014).
- 17.Organization, W. H. WHO meeting report of a technical expert consultation: non-inferiority analysis of Xpert MTB/RIF Ultra compared to Xpert MTB/RIF. (World Health Organization, 2017).
- 18.Hain Lifescience. GenoType MTBDRsl VER 2.0 Instructions for Use. (2015).
- 19.Motsoaledi, A. Health Department of the Republic of South Africa (2014).
- 20.Cepheid. Journey Inside the Cepheid GeneXpert® Cartridge - 3D Animation (2019).
- 21.Raja S, et al. Technology for automated, rapid, and quantitative PCR or reverse transcription-PCR clinical testing. 2005;51:882–890. doi: 10.1373/clinchem.2004.046474. [DOI] [PubMed] [Google Scholar]
- 22.Theron, G. In Molecular Microbiology: Diagnostic Principles and Practice (ed. David H. Persing et al.) Ch. 40, (ASM Press, 2016).
- 23.Chakravorty S, Helb D, Burday M, Connell N, Alland D. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. Journal of microbiological methods. 2007;69:330–339. doi: 10.1016/j.mimet.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Derendinger B, et al. Widespread use of incorrect PCR ramp rate negatively impacts multidrug-resistant tuberculosis diagnosis (MTBDR plus). 2018;8:3206. doi: 10.1038/s41598-018-21458-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hain Lifescience. FluoroType® MTBDR VER 2.0 Instructions for Use. (2019).
- 26.Mishra, H. et al. Xpert MTB/RIF Ultra and Xpert MTB/RIF for diagnosis of tuberculosis in an HIV-endemic setting with a high burden of previous tuberculosis: a two-cohort diagnostic accuracy study. Lancet Respiratory Medicine, 10.1016/S2213-2600(19)30370-4 (2020). [DOI] [PubMed]
- 27.Mao X, et al. Diagnosis of drug resistance to fluoroquinolones, amikacin, capreomycin, kanamycin and ethambutol with Genotype MTBDRsl assay: a meta-analysis. Annals of Clinical & Laboratory Science. 2015;45:533–544. [PubMed] [Google Scholar]
- 28.Theron, G. et al. The diagnostic accuracy of the GenoType (®) MTBDRsl assay for the detection of resistance to second-line anti-tuberculosis drugs. Cochrane Database of Systematic Reviews10 (2014). [DOI] [PMC free article] [PubMed]
- 29.Tomasicchio, M. et al. The diagnostic accuracy of the MTBDRplus and MTBDRsl assays for drug-resistant TB detection when performed on sputum and culture isolates. Scientific reports6 (2016). [DOI] [PMC free article] [PubMed]
- 30.World Health Organization. WHO treatment guidelines for drug-resistant tuberculosis. (2016).
- 31.Blakemore R, et al. Evaluation of the analytical performance of the Xpert MTB/RIF assay. Journal of clinical microbiology. 2010;48:2495–2501. doi: 10.1128/JCM.00128-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Theron G, et al. Xpert MTB/RIF results in patients with previous tuberculosis: can we distinguish true from false positive results? Clinical Infectious Diseases. 2016;62:995–1001. doi: 10.1093/cid/civ1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Theron, G. et al. False positive Xpert MTB/RIF results in re-tested patients with previous tuberculosis: frequency, profile, and prospective clinical outcomes. Journal of clinical microbiology, JCM. 01696–01617 (2018). [DOI] [PMC free article] [PubMed]
- 34.World Health Organization. The use of molecular line probe assay for the detection of resistance to isoniazid and rifampicin: policy update. (2016).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on request.