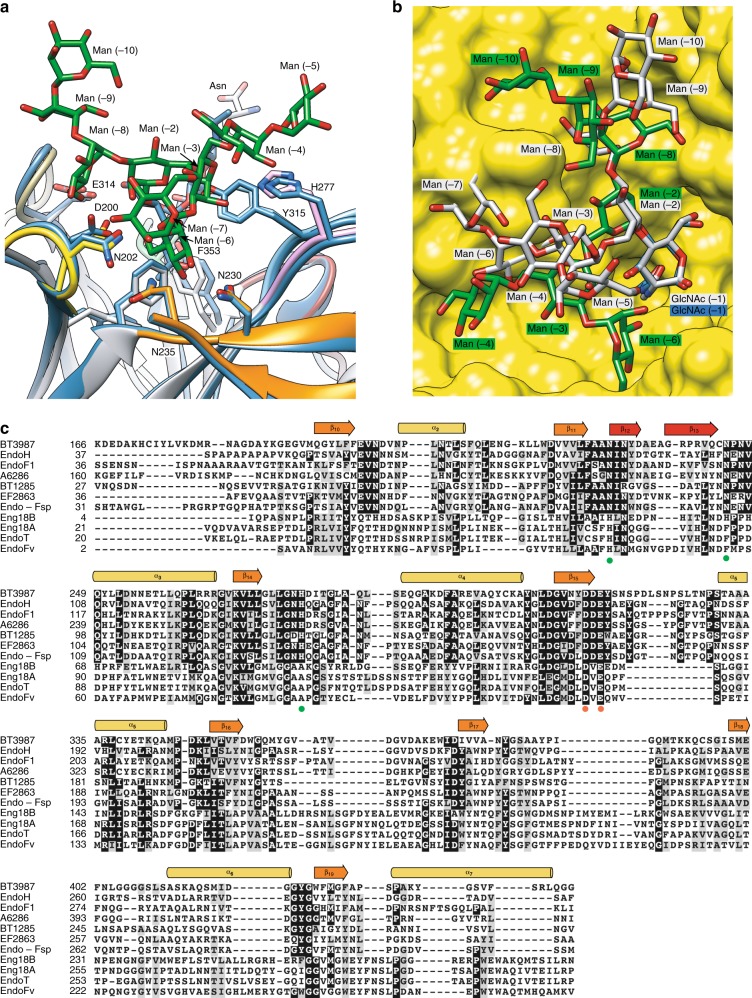
Fig. 5. Structural basis of EndoBT-3987 specificity for HM-type N-glycans.
a Structural comparison of EndoBT-3987D312A/E314L-Man9GlcNAc2Asn in gray and EndoH (PDB CODE 2EBN) in blue. The residues involved in substrate recognition are colored by loops: loop 1 (yellow), loop 2 (orange), and loop 3 (pinked). b Structural comparison of the HM-type glycan conformation in the active site of EndoBT-3987WT-Man9GlcNAc-2 (gray) and EndoS2 (PDB CODE 6MDV) (green) on surface representation of binding site of EndoS2 (yellow). c Structure weighted sequence alignment of BT3987 with GH18 ENGases family with characterized endo-N-acetyl-β-D-glucosaminidase activity against HM-type N-glycans and inactive against CT-type N-glycans. Comparison of BT3987 from B. thetaiotaomicron VPI-5482 (Q8A0N4, Uniprot code), EndoH from Streptomyces plicatus (P04067, Uniprot code), EndoF1 from Elizabethkingia meningoseptica (P36911, Uniprot code), A6286 from Prevotella melaninogenica (D9RSV7, Uniprot code), BT1285 from B. thetaiotaomicron VPI-5482 (Q8A889, Uniprot code), EF2863 from Enterococcus faecalis (Q830C5, Uniprot code), Endo-Fsp from Flavobacterium sp. (P80036, Uniprot code), Eng18B from Hypocrea atroviride IMI 206040 (G9P8KO, Uniprot code), Eng18A from Hypocrea atroviride IMI 206040 (G9NR36, Uniprot code), EndoT from Hypocrea jecorina (C4RA89, Uniprot code), and EndoFv from Flammulina velutipes (D1GA49, Uniprot code). The catalytic residues are marked with red dots and the key residues that interact with Man9 are marked with green dots.