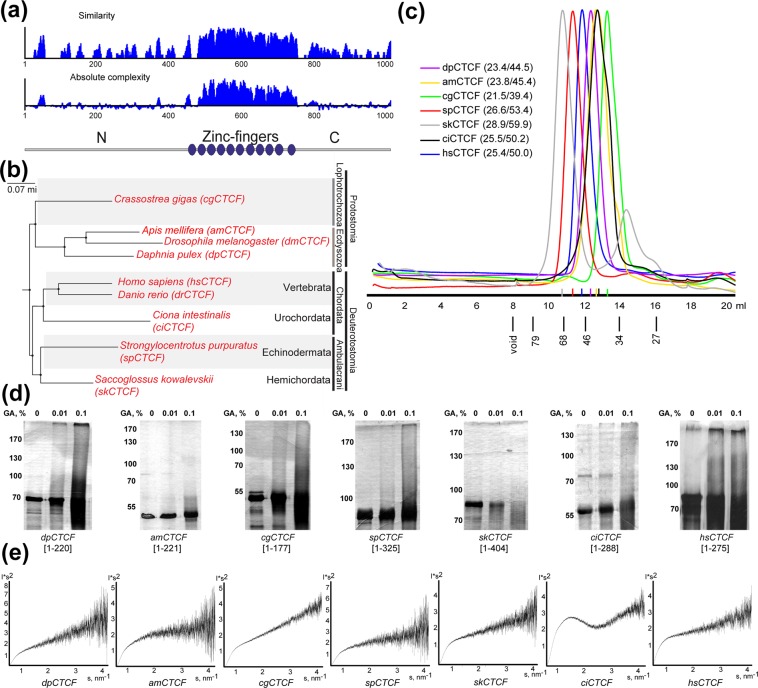
Figure 2.
(a) Summary of multiple sequence alignment of CTCF proteins used in this study. The similarity is a score of how similar each amino acid or groups of amino acids are across the whole alignment. The absolute complexity is the average of the pairwise alignment scores using the substitution matrix chosen in the alignment setup. Overall domain structure of CTCF proteins is shown below. (b) Positions of species selected for this study on the phylogenetic tree of metazoans (adapted from38). (c) Superdex S200 size-exclusion chromatography of CTCF NTDs (without Thioredoxin). Elution volumes of proteins with known Rs values (Å) are shown. Calculated Rs values for NTDs are shown in brackets (globular/unfolded monomer). (d) Cross-linking of Thioredoxin-tagged NTDs using increasing concentrations of glutaraldehyde (GA). (e) Kratky plot (I*s2 vs s) of SAXS data derived for CTCF NTDs. The bell-shaped curve suggests that polypeptide is folded, whereas the logarithmic shape is a sign of random coil conformation.