Fig. 4.
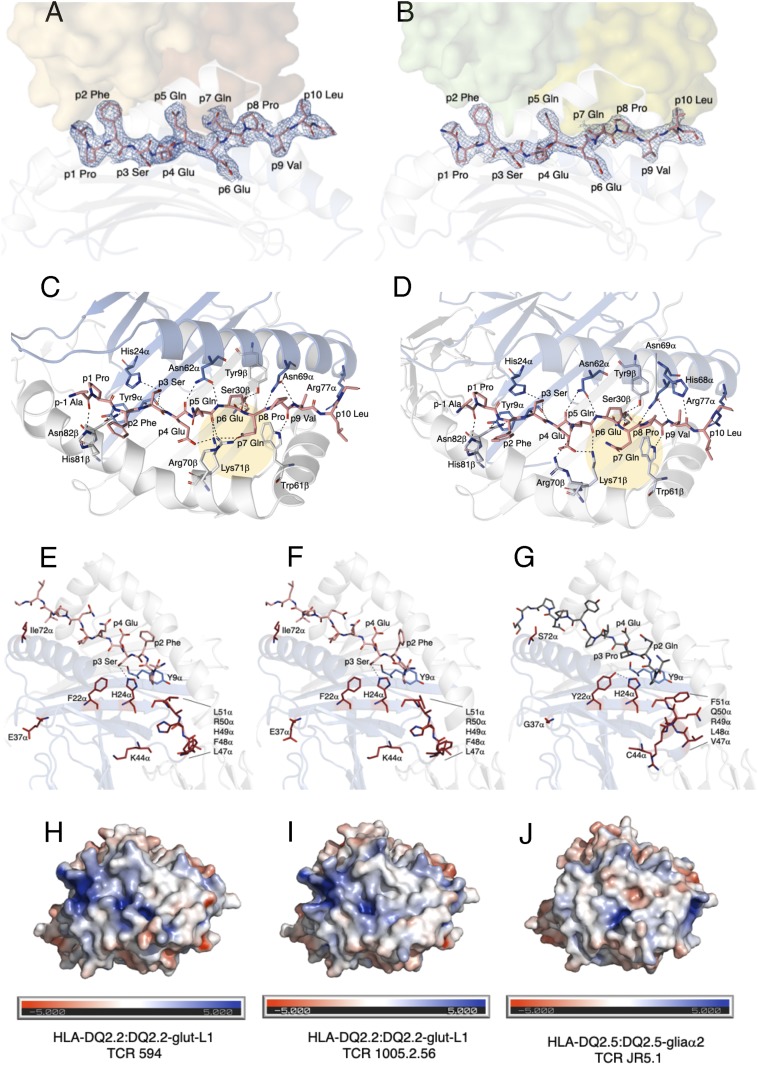
Comparison of HLA-DQ2.2-glut-L1 and HLA-DQ2.5 in complexed with gluten epitopes. The DQ2.2-glut-L1 peptide is bound in the peptide-binding groove, with carbon colored in salmon, nitrogen colored in blue, and oxygen colored in red. The TCR 594 (A) and TCR 1002.56 (B) docked on top of the HLA-DQ2.2:DQ2.2-glut-L1 are shown as solid surface, and each of the TCR αβ pair were colored in brown/orange and green/lime, respectively. The peptide’s 2Fo-Fc electron density map is shown in blue and contoured to 1 σ. Hydrogen bond interactions between HLA-DQ2.2 and the DQ2.2-glut-L1 epitope in the TCR 594:HLA-DQ2.2:DQ2.2-glut-L1 ternary complex (C) and in the TCR 1005.2.56:HLA-DQ2.2:DQ2.2.-glut-L1 ternary complex (D) are shown. Comparison of the DQ2.2-glut-L1 peptide bound to HLA-DQ2.2 in the two ternary complexes solved showed minor movement in P7-Gln residues, highlighted in yellow circle. Polymorphic residues on the ectodomain of HLA-DQ differing between HLA-DQ2.2 (E and F) and HLA-DQ2.5 (G) are represented in stick and colored in fire-brick. The His24α interacts with P3-Ser in DQ2.2-glut-L1 (E and F) but with Tyr22α in HLA-DQ2.5 (G). The solvent-accessible electrostatic potential was calculated for HLA-DQ2.2:DQ2.2-glut-L1 in complexed with TCR 594 (H) or TCR 1005.2.56 (I) and HLA-DQ2.5:DQ2.5-glia-α-2 (J). Electrostatic calculations were performed using APBS (±5 kT/e).