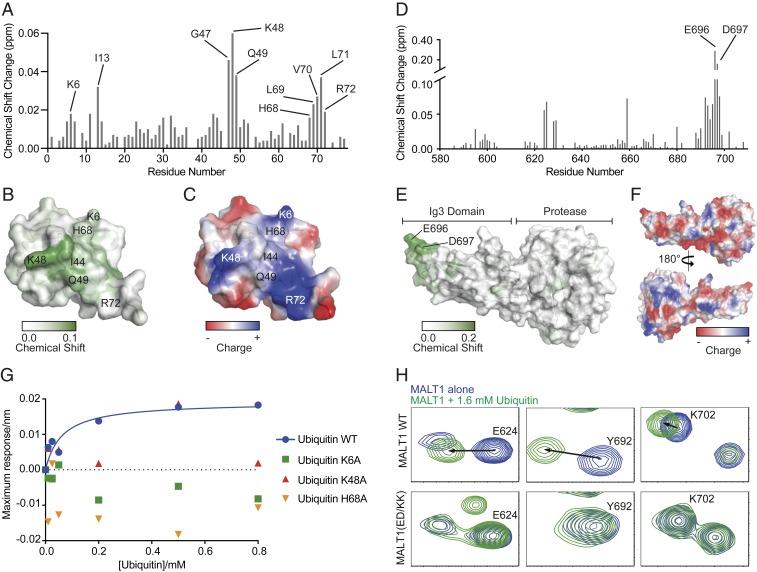
Fig. 1.
Monomeric ubiquitin binds to the Ig3 domain of MALT1. (A) Backbone amide minimal shifts seen for 15N-labeled human ubiquitin (residues 1–76) upon mixing with an over fivefold molar excess of purified MALT1 (residues 339–719). (B) Mapping of the minimal shift backbone amide NMR data onto a surface representation of ubiquitin with significantly perturbed residues (shift >0.01 ppm) colored with a gradient from white to green. Residues for which no minimal shift data were obtained are also shown in white. (C) An electrostatic surface representation of ubiquitin shown in the same orientation as B with areas of significant positive charge indicated in blue, and areas with negative charge indicated in red. Images in B and C were prepared using PyMOL. (D and E) As in A and B, showing backbone amide shifts seen for 15N-labeled human MALT1 (residues 339–719) upon addition of an over 10-fold molar excess of human ubiquitin (residues 1–76). Data in D are only shown for amino acids constituting the Ig3 domain (580–709) as no significant changes were seen for backbone amide signals from residues in the MALT1 protease domain. (F) Surface charge representation of the MALT1 Ig3 and protease domains (positive charges are indicated in blue, and negative charges are indicated in red), highlighting the negatively charged surface around the region affected by ubiquitin binding. (G) MALT1 (residues 339–719) maximum binding response as a function of increasing concentrations of ubiquitin (residues 1–76) WT (blue filled circles), K6A (green filled squares), K48A (red filled upward triangles), and H68A (orange filled downward triangles). MALT1 WT binds to ubiquitin WT according to a one-site saturation-binding model with a dissociation constant (KD) of ∼62.0 μM. No binding is detected between MALT1 WT and ubiquitin variants K6A, K48A, and H68A. (H) Highlighted backbone amide peaks from the 15N-1H TROSY spectra for E624, Y692, and K702, which show substantial shifts in MALT1 WT but not in MALT1(ED/KK) upon addition of ubiquitin.