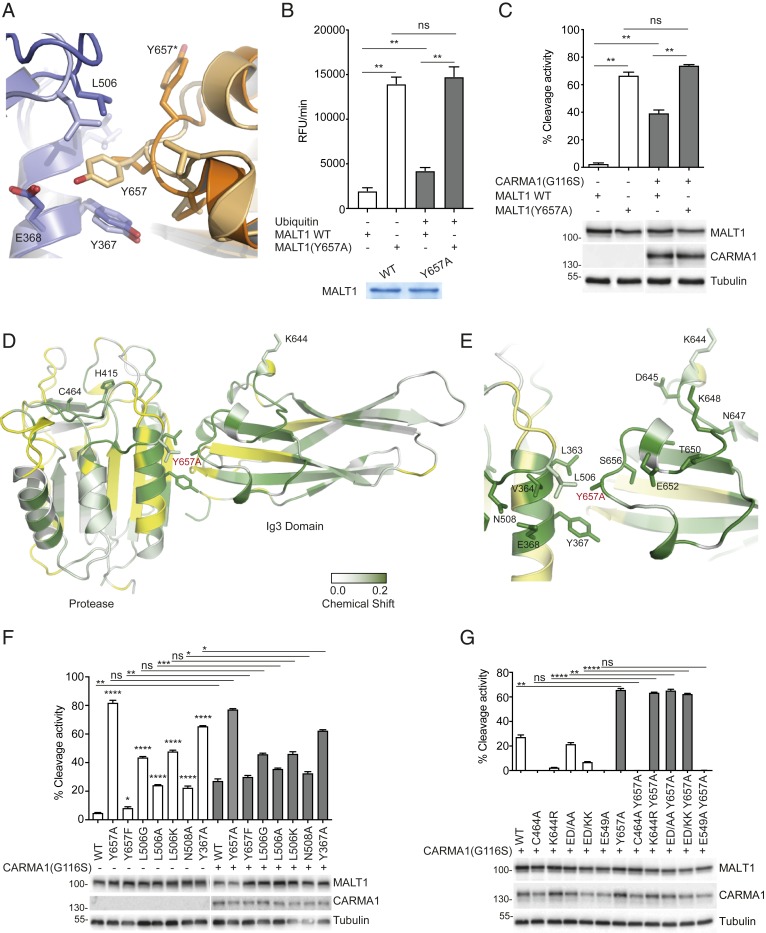
Fig. 4.
Mutation of Y657 induces coordinated conformational changes in the loop connecting K644 to Y657 and the active site. (A) Overlay of the MALT1 Ig3-protease interface from reported crystal structures, which shows a major conformational switch of Y657 in the presence (light orange/light blue [PDB: 3UO8]) and the absence (dark orange/dark blue [PDB: 3V55]) of the active site peptide inhibitor z-VRPR-fmk. Blue and orange colors represent the protease and Ig3 domains, respectively. (B) In vitro cleavage assay comparing the capacity of ubiquitin (0.1 mM) to activate WT MALT1 or a MALT1 Y657A mutant (residues 199–824, 0.6 µM). Protein amount was controlled by Coomassie-blue staining as indicated. (C) Flow cytometric assessment of MALT1-dependent FRET reporter cleavage in 293T cells. The reporter was coexpressed with the indicated MALT1 constructs together with oncogenic CARMA1(G116S). Protein expression was controlled by Western blotting as indicated, and positions of molecular weight markers (in kDa) are indicated (C). (D and E) Backbone amide chemical shift changes induced by the Y657A mutation in MALT1 and mapped onto the reported structure of the Ig3-protease domains. Residues with significantly perturbed NMR signals (shift > 0.01 ppm) are colored with a gradient from white to green. Residues for which no minimal shift data were obtained are shown in yellow. Figure prepared using PyMOL. Residues with clearly shifted signals located at the Ig3-protease interface are highlighted (E). (F and G) Cleavage activity of the indicated MALT1 constructs was assessed as in C. Data are representative of two (C, F, and G) or three (B) experiments. Bars represent means ± SD; ns, nonsignificant, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.