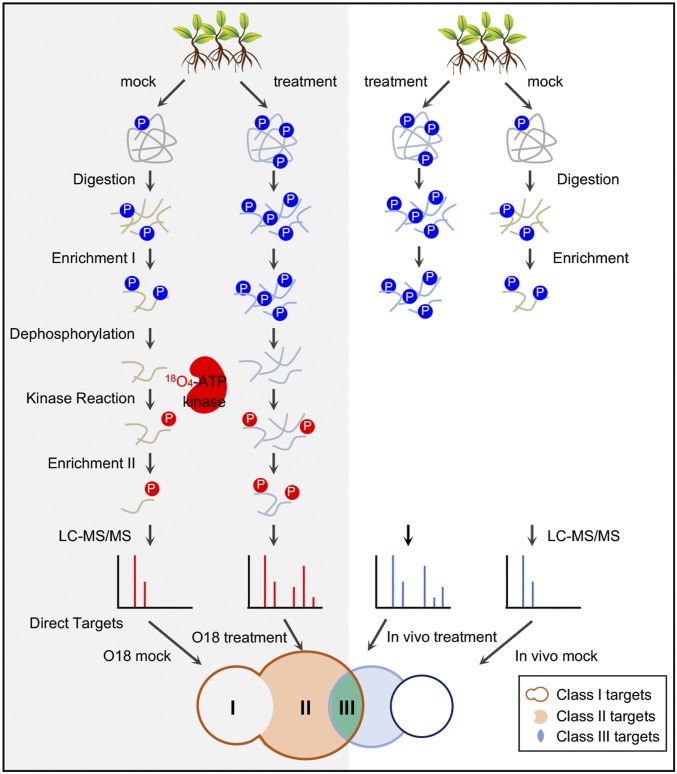
Fig. 1.
The KALIP 2.0 workflow for identifying kinase substrate proteins. Protein extracts from mock- and stress-treated seedlings were digested with Lys-C, and the resulting long phosphopeptides (blue) were enriched by polyMAC. The phosphopeptides were dephosphorylated and incubated with a recombinant kinase and γ-18O4-ATP, and 18O-phosphate-labeled peptides (red) were enriched again through polyMAC and analyzed by LC-MS/MS. Proteins with heavy (18O) phosphopeptides identified from all samples were classified as class I substrates (red circle). The phosphoproteins with heavy phosphosites significantly enriched in the stress-activated samples (FDR < 0.05) were classified as class II substrates (orange). Phosphoproteins significantly enriched in stress-treated samples in both the in vitro kinase reaction and in vivo kinase-dependent phosphoproteomic comparison were classified as class III substrates (green).