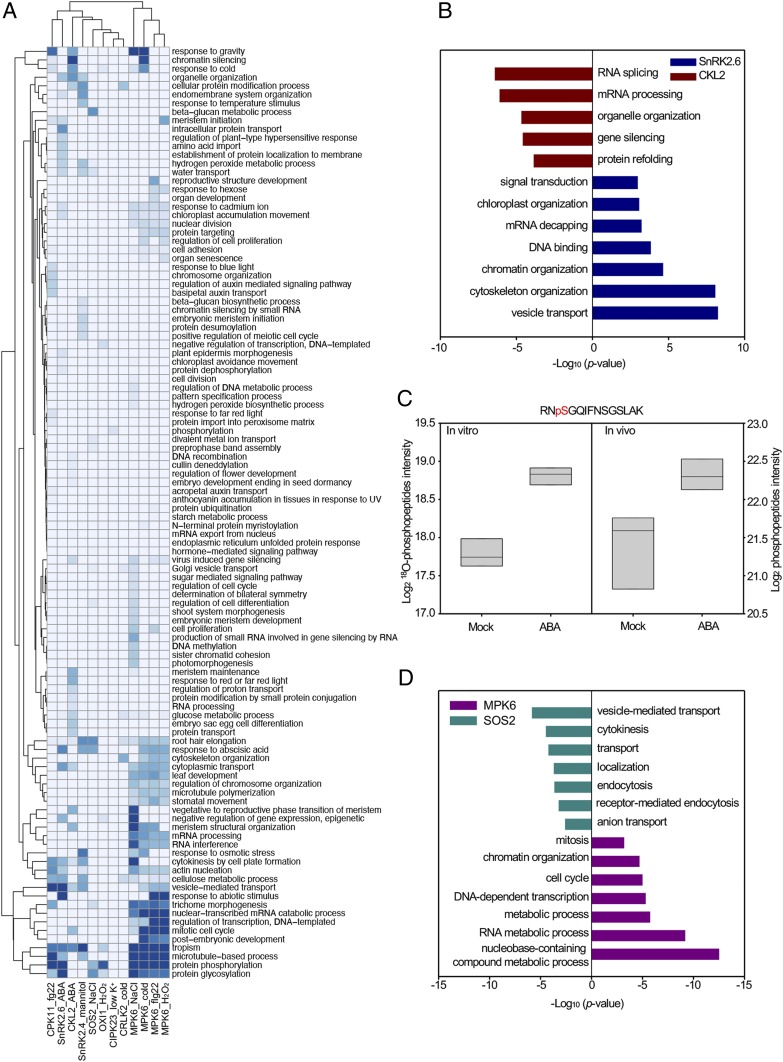
Fig. 4.
Proteomic signature of the putative substrates (unique proteins) identified from 12 protein kinase/treatment combinations. (A) GO analysis of the putative substrates (unique proteins) identified from 12 protein kinase/treatment combinations. (B) Bar chart highlighting enriched categories of GO biological function in putative SnRK2.6 and CKL2 substrates identified from ABA-treated seedlings using Fisher’s exact test (Benjamini–Hochberg FDR < 0.05). (C) CKL2 is a class III putative substrate of SnRK2.6. The phosphopeptide RNpSGQIFNSGSLAK was identified both in vitro and in vivo, and its level was significantly increased in ABA-treated samples in both the in vitro and in vivo comparisons (two-sample t test, FDR < 0.05). (D) Dark cyan and pink bars representing significant GO categories in putative SOS2 and MPK6 substrates identified from NaCl-treated seedlings (Benjamini–Hochberg FDR < 0.05).