Significance
Skeletal muscle cells are unique among human cells because they contain hundreds of nuclei distributed over large distances within a shared cytoplasm. This unusual organization raises questions about how gene expression is coordinated between nuclei and how parts of the muscle cell can become functionally distinct. Since transcription factors and other nuclear proteins control gene expression, we investigated how far nuclear proteins can travel between nuclei within a single muscle cell. We found that we could predict how far a nuclear protein would travel based on its size, nuclear import speed, and ability to diffuse across the nuclear pore. These findings can help explain how muscle cells are organized and provide design parameters for engineering gene therapies in skeletal muscle.
Keywords: skeletal muscle, myonuclear domain, nuclear transport, mathematical model
Abstract
Skeletal muscle cells contain hundreds of myonuclei within a shared cytoplasm, presenting unique challenges for regulating gene expression. Certain transcriptional programs (e.g., postsynaptic machinery) are segregated to specialized domains, while others (e.g., contractile proteins) do not show spatial confinement. Furthermore, local stimuli, such as denervation, can induce transcriptional responses that are propagated along the muscle cells. Regulated transport of nuclear proteins (e.g., transcription factors) between myonuclei represents a potential mechanism for coordinating gene expression. However, the principles underlying the transport of nuclear proteins within multinucleated cells remain poorly defined. Here we used a mosaic transfection model to create myotubes that contained exactly one myonucleus expressing a fluorescent nuclear reporter and monitored its distribution among all myonuclei. We found that the transport properties of these model nuclear proteins in myotubes depended on molecular weight and nuclear import rate, as well as on myotube width. Interestingly, muscle hypertrophy increased the transport of high molecular weight nuclear proteins, while atrophy restricted the transport of smaller nuclear proteins. We have developed a mathematical model of nuclear protein transport within a myotube that recapitulates the results of our in vitro experiments. To test the relevance to nuclear proteins expressed in skeletal muscle, we studied the transport of two transcription factors—aryl hydrocarbon receptor nuclear translocator and sine oculis homeobox 1—and found that their distributions were similar to the reporter proteins with corresponding molecular weights. Together, these results define a set of variables that can be used to predict the spatial distributions of nuclear proteins within a myotube.
Skeletal muscle plays a critical role in human health by enabling movement and regulating metabolic activity. Myofibers, the primary cells in skeletal muscle, form from the fusion of skeletal muscle progenitors, known as myoblasts. Myofibers contain numerous myonuclei distributed over large distances, spanning the entire length of a muscle. Although they share a common cytoplasm, myonuclei within a single myofiber can exhibit significant variation in gene expression (1), leading to questions of how gene expression is coordinated and how local transcriptional responses to stimuli are propagated within myofibers.
It has been proposed that each myonucleus in a myofiber produces proteins for a region of cytoplasm surrounding it, called the myonuclear domain (2). However, myonuclear domains are conceptual; in reality, proteins produced by one myonucleus are likely transported over different distances along the length of the myofiber, depending on their specific characteristics (e.g., molecular weight, subcellular localization, degradation rate). Protein transport along a myofiber may provide a mechanism for a small number of myonuclei in one part of a muscle myofiber to affect the function of the entire cell. This has been proposed as a paradigm for cell-based therapies in muscle, for example, to treat muscular dystrophy (3). However, the characteristics that affect the transport of proteins from one myonucleus to other regions of the myofiber remain largely unknown.
The transport of nuclear proteins (e.g., transcription factors) within myofibers is of particular interest because of their potential to coordinate gene expression. For example, in the case of muscle innervation, the ETS family transcription factor GA-binding protein localizes specifically to postsynaptic myonuclei at the neuromuscular junction (4) to initiate the expression of acetylcholine receptor subunits α (5) and ε (6), which are not expressed in other regions of the myofiber. Furthermore, throughout the myofiber, there are significant variations in the levels of certain transcription factors, including NFAT5 (7), NFATc1 (8), MyoD (9), and myostatin (10), which regulate muscle growth and differentiation (11). This variation suggests that regions of a myofiber may respond to a localized signal in a spatially coordinated fashion. How nuclear proteins that are produced in a shared cytoplasm become sequestered to specific myonuclei remains unclear, however.
The transport of proteins from the cytoplasm into myonuclei relies largely on facilitated import pathways; however, it is not known how far nuclear proteins produced by a single myonucleus can travel before entering a neighboring myonucleus and how the characteristics of the nuclear protein affect this distance. To address these limitations, we used a mosaic transfection approach to produce myotubes containing exactly one myonucleus expressing a fluorescent nuclear reporter. We quantified the relative amount of this reporter that entered myonuclei neighboring the transfected myonucleus and compared the transport of different closely related nuclear proteins. These data were used together with a mathematical model to identify key characteristics affecting the propagation of nuclear proteins within myotubes.
Results
A Mosaic Transfection Model for Quantifying Nuclear Protein Propagation.
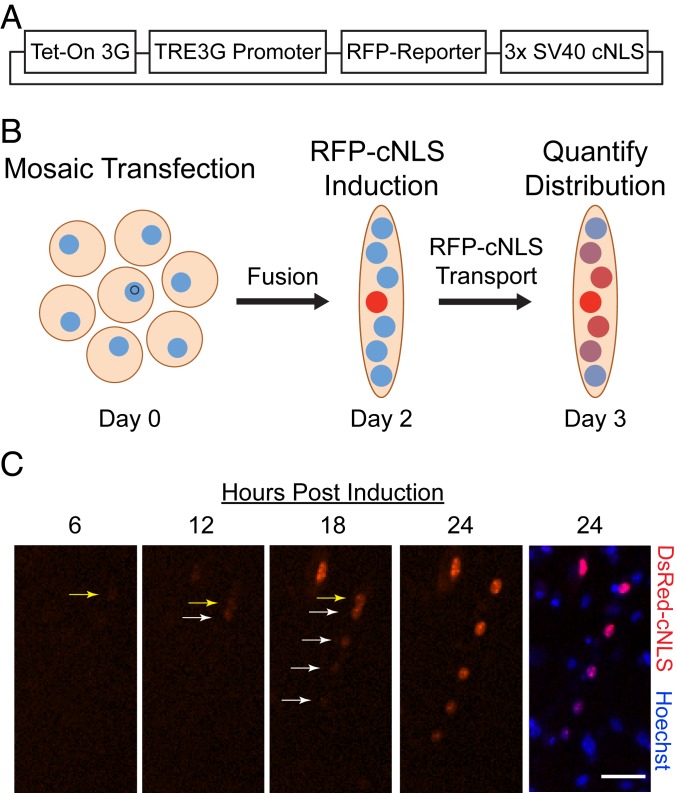
When myoblasts differentiate and fuse in vitro, the resulting multinucleated and postmitotic cells are called myotubes. To measure the distance that nuclear proteins expressed by one myonucleus can travel before entering neighboring myonuclei, we generated myotubes containing exactly one myonucleus expressing a fluorescent nuclear reporter. Tetracycline-inducible expression vectors were used to control the expression of these reporters, which consisted of a red fluorescent protein (RFP) fused with three copies of the simian virus 40 large T antigen classical nuclear localization sequence (3× SV40 cNLS), termed RFP-cNLS (Fig. 1A). A cNLS sequence was chosen because it is the most common type of NLS among nuclear proteins, and the SV40 cNLS is among the most well-characterized of these sequences (12). Three RFP species—mCherry (28.8 kDa) (13), tdTomato (55 kDa) (13), and DsRed-Express2 (DsRed; a 27.6-kDa monomer that forms a 110-kDa tetramer) (14)—were used because they have different diffusion coefficients that are inversely related to molecular weight while being closely related in terms of chemical characteristics. On induction with doxycycline, all three RFPs exhibited bright red fluorescence and were localized to the nuclei of transfected primary mouse myoblasts (SI Appendix, Fig. S1A). No RFP expression was observed in the absence of doxycycline.
Fig. 1.
A mosaic transfection model for quantifying nuclear protein propagation. (A) RFPs of varying molecular weight (RFP-reporter) were fused with 3× SV40 cNLS in a tetracycline-inducible vector. (B) Primary mouse myoblasts were transfected with an RFP reporter plasmid at 1 h after seeding, resulting in a sparse mosaic transfection (see also SI Appendix, Fig. S1). Myoblasts were then differentiated for 2 d without doxycycline. Following myotube fusion, doxycycline was added to the culture medium for 24 h to induce expression of the RFP-cNLS fusion protein, and the RFP distributions within individual myotubes were quantified by epifluorescence microscopy. (C) Time-lapse microscopy of a Hoechst-stained myotube following induction with doxycycline showed that the myotube contained a single transfected myonucleus expressing the DsRed-cNLS fusion protein (see also Movies S1 to S6). Initially, the RFP-cNLS fusion protein was imported into the transfected myonucleus (yellow arrow), but after 12 h, the reporter protein was also imported into neighboring myonuclei (white arrows). This process continued for 24 h after doxycycline induction, resulting in a gradient of myonuclear RFP fluorescence, which we refer to as the propagation profile for that protein. (Scale bar: 50 µm.)
To create the mosaic model, primary mouse myoblasts were transfected with the RFP-cNLS inducible-expression vectors to generate myoblast cultures in which <∼2.5% of the cells were positively transfected (SI Appendix, Fig. S1B). The myoblasts were then differentiated for 48 h to form myotubes (Fig. 1B). As a result of the low transfection rate, <5% of the myotubes (containing ∼15 myonuclei) were predicted to contain more than one transfected myonucleus. After myotube fusion, doxycycline was added to the differentiation medium to induce expression of the RFP-cNLS protein, and transport of the reporter was monitored by time-lapse epifluorescence microscopy (Movies S1–S6). Initially, the RFP-cNLS fusion protein was imported mainly into the transfected myonucleus, but after 10 to 12 h, the reporter protein was also observed inside neighboring myonuclei (Fig. 1C). The spread of the fluorescent nuclear reporter continued for at least 24 h after induction, resulting in a gradient of myonuclear RFP fluorescence within the myotube (Fig. 1C). By contrast, transfected myotubes containing single nuclei expressing either an untargeted green fluorescent protein (CopGFP; 24.7 kDa) or one of two fluorescent reporters targeted to mitochondria (AcGFP1-MLS, 26.9 kDa; DsRed2-MLS, 103.2 kDa) exhibited limited or absent gradients of signal (SI Appendix, Fig. S2). The results from the untargeted fluorescent protein suggest that diffusion alone, without organelle localization, is insufficient to create gradients within myotubes, while the results of the two fluorescent proteins targeted to the mitochondria imply that organelle dynamics (e.g., transport, fusion and fission) can partially resolve gradients that are initially created by organelle targeting, irrespective of the diffusion coefficient of the fluorescent protein (15, 16).
These experiments provide a quantitative measure of the spatial distribution of nuclear proteins along myotubes. Nuclear proteins have the potential per se to affect gene expression (e.g., transcription factors); therefore, their limited transport could lead to localized transcriptional domains, as has been observed within myofibers in vivo (1). Since transcription factors transmit gene expression signals within cells, we have chosen to use the term “propagation” to describe the distance that these signals travel within myotubes.
Molecular Weight Affects the Propagation of Nuclear Proteins.
Since nuclear proteins exist over a wide range of molecular weights, we examined how the molecular weights of the RFP-cNLS fusion proteins affected their propagation. Three closely related RFPs were used— mCherry, tdTomato, and DsRed-Express2—which represent monomer, dimer and tetramer variants, respectively, of the RFPs isolated from Discosoma sp. (13, 14). This strategy allowed us to study how molecular weight affects the propagation of nuclear proteins while maintaining other chemical properties.
All RFP-cNLS fusion proteins propagated from the transfected myonucleus and localized partly within neighboring myonuclei (Fig. 2A). Within myonuclei, the RFPs partially colocalized with the nucleolus (SI Appendix, Fig. S3), with DsRed-cNLS observed as bright punctae. By contrast, the mCherry and tdTomato constructs presented as more diffuse fluorescence (Fig. 2A). It is unlikely that the punctae are the result of DsRed forming higher-order structures, because this fluorophore was designed to prevent such aggregates (14) and the punctae were not observed outside the nuclei. Localization of RFPs to a subnuclear compartment is unlikely to affect their propagation because cytoplasmic diffusion and facilitated nuclear import should be unaffected.
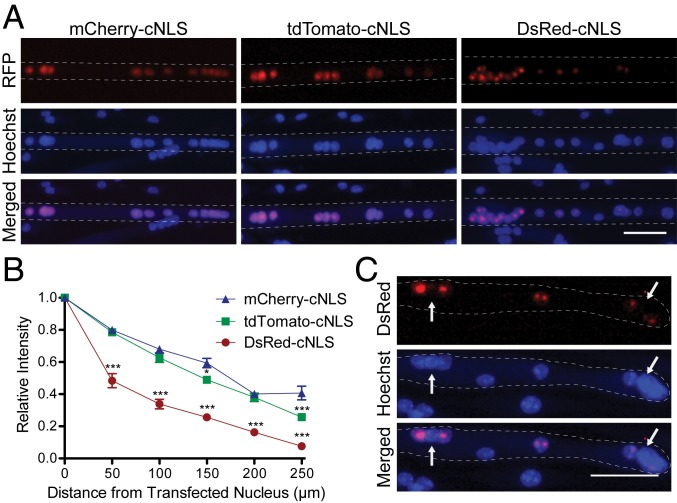
Fig. 2.
Myonuclear propagation is affected by the molecular weight of the RFP-cNLS fusion protein. Myoblasts were transfected with inducible expression vectors for cNLS-RFP fusion proteins of varying molecular weight (mCherry, 28.8 kDa; tdTomato, 55 kDa; DsRed, which forms a 110-kDa tetramer) and then differentiated for 2 d. (A) After 24 h of doxycycline induction, the distributions of RFP fluorescence within Hoechst-stained myotubes were visualized by epifluorescence microscopy. (B) RFP fluorescence was quantified by measuring the positions and average intensity of myonuclei within individual transfected myotubes. Distances and fluorescence intensities were normalized to the brightest myonucleus within each myotube (assumed to be the transfected nucleus). Data are binned by distance (bin size, 50 µm) and are represented as mean ± SEM (two-way analysis of variance [ANOVA] with Bonferroni posttest). (C) A small portion of the myotubes expressing the largest molecular weight RFP-cNLS fusion protein (DsRed) contained individual myonuclei that did not import the reporter protein as expected (arrows). These myotubes were excluded from the analysis in B. (Scale bars: 50 µm in A and C.) *P < 0.05; ***P < 0.001.
To measure propagation, RFP fluorescence and spatial position were recorded for all myonuclei within individual myotubes. The distances and average fluorescence intensities were then normalized to the brightest myonucleus within each myotube. It was assumed that the brightest myonucleus was the one producing the RFP-cNLS fusion protein, as indicated by time-lapse microscopy (Movies S1 to S6), which was referred to as the transfected nucleus. Fluorescence measurements from all other nuclei were binned based on their distance from the transfected nucleus, producing a trace of relative fluorescence intensity versus distance from the transfected nucleus (Fig. 2B). This analysis allowed for a simplified comparison of nuclear protein propagation across various conditions.
The molecular weight of the RFP-cNLS constructs was inversely correlated to their propagation (Fig. 2B). For example, at a distance 100 to 150 µm from nuclei transfected with the mCherry-cNLS construct, myonuclei had ∼60% the fluorescence intensity of the transfected nucleus. By comparison, myonuclei at the same distances had only 49% and 25% the fluorescence intensity for the tdTomato-cNLS and DsRed-cNLS constructs, respectively (Fig. 2B). Importantly, there were no significant differences in the distributions of the RFP-cNLS transcripts (SI Appendix, Fig. S4), indicating that the differences in construct propagation (Fig. 2B) were the result of the transport characteristics of the proteins rather than those of the transcripts. Furthermore, the average intensity of the Hoechst counterstain signal did not vary among RFP isoforms or along the length of a myotube, indicating that the RFP gradients observed did not originate from gradients in the spatial distributions of the nuclei (SI Appendix, Fig. S5).
To exclude the possibility that artifacts derived from the transient transfection method affected the results, we reproduced the mosaic model using lentiviral transduction. Myoblasts containing integrated copies of each of the inducible RFP-cNLS inserts were selected by fluorescence-activated cell sorting. Sorted RFP-cNLS+ myoblasts were mixed with wild-type myoblasts at a 1:50 ratio, followed by differentiation and induction of RFP expression. Propagation of nuclear RFPs closely matched that observed following transient transfection (SI Appendix, Fig. S6). Sorted RFP-cNLS+ myoblast populations also enabled nuclear tracking via incorporation of the thymidine analog 5-ethynyl-2′-deoxyuridine (EdU). RFP-cNLS+ myoblasts were cultured in the presence of EdU for 24 h prior to mixing and differentiation. Subsequent visualization of EdU+ nuclei at the experimental endpoint showed that in each case, the brightest nucleus or its immediate neighbor was derived from the sorted population and therefore contained the RFP-cNLS expression cassette (SI Appendix, Fig. S7).
Interestingly, the largest difference among the three constructs occurred in myonuclei that were close (0 to 50 µm) to the transfected nucleus. At these distances, myonuclei importing mCherry-cNLS and tdTomato-cNLS both contained ∼80% the RFP signal of the transfected nucleus, while the nuclei importing the DsRed-cNLS construct contained ∼50% (Fig. 2B). One possible explanation for this finding is that the smaller RFP-cNLS proteins are able to diffuse bidirectionally across the nuclear pore complex (NPC), which might allow the smaller proteins to “escape” the transfected nucleus and enter neighboring myonuclei, thereby creating a broader propagation profile (Fig. 2B). In fact, proteins as large as 60 kDa have been reported to diffuse across the NPC (17), making it likely that diffusion across the NPC plays a role in the propagation of mCherry-cNLS and tdTomato-cNLS proteins.
The hypothesis that the smaller RFP-cNLS constructs could diffuse across the NPC is supported by our observations of “nuclear skipping.” A small proportion of the myotubes transfected with the DsRed-cNLS construct contained one or more nuclei that appeared incapable of importing the 110-kDa tetramer (Fig. 2C). This result suggests that not all myonuclei in a myotube are equally competent at importing proteins through the classical nuclear import pathway, as has been shown previously (18, 19). By contrast, nuclear skipping was not observed in myotubes transfected with the smaller constructs. This suggests that these constructs were able to enter all myonuclei, possibly by diffusion across the NPC, even if the myonuclei were not competent to import proteins through facilitated nuclear import. Myotubes with nuclear skipping were excluded from our analysis of propagation.
Facilitated Nuclear Import Limits Propagation of Nuclear Proteins.
To examine the mechanisms regulating the propagation of nuclear proteins, we interfered with the classical nuclear import pathway. Importazole is a small-molecule inhibitor of the RAN-GTP/importin-β association that has been shown to inhibit the nuclear import of cNLS proteins in a variety of contexts (20). Therefore, importazole was used to test how the classical import pathway regulates the propagation of nuclear proteins.
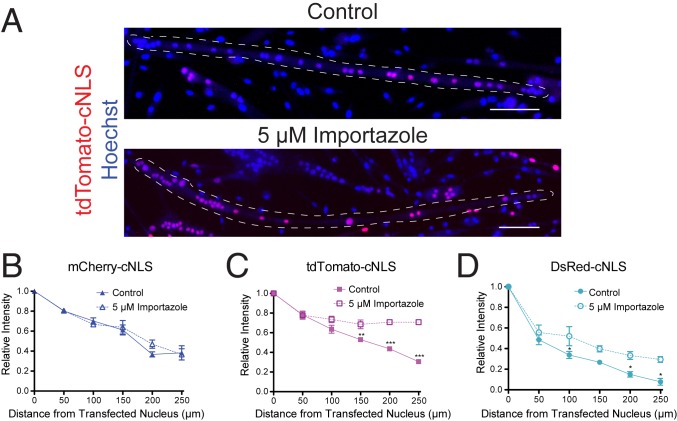
To minimize the cytotoxic effects of inhibiting nuclear import, 5 µM importazole was added to myotubes only during the final 24 h of culture, when doxycycline was added to induce RFP-cNLS expression. Importazole is capable of completely inhibiting the classical import pathway at concentrations >40 µM (20); however, we found concentrations >20 µM to be cytotoxic to myotubes. Therefore, we used a low concentration of importazole (5 µM) to slow nuclear import while still allowing the RFP-cNLS fusion proteins to localize to myonuclei (Fig. 3A). Even at this low concentration, importazole effected the propagation of some RFP-cNLS proteins; for example, whereas tdTomato-cNLS was normally found only in one region of a myotube, importazole treatment resulted in myotubes that contained tdTomato-cNLS distributed throughout the whole cell (Fig. 3A). Interestingly, the distribution of the smallest RFP-cNLS fusion protein (mCherry-cNLS) was not affected by importazole treatment (Fig. 3B). This suggests that the propagation of small nuclear proteins may be dictated primarily by passive diffusion across the NPC rather than by active nuclear import. By contrast, the propagation of both the tdTomato-cNLS and DsRed-cNLS fusion proteins was extended by importazole treatment (Fig. 3 C and D). These data show that the classical import pathway limits the myonuclear propagation of larger proteins by sequestering them into neighboring myonuclei, creating a localized gradient.
Fig. 3.
Propagation of large RFP-cNLS fusion proteins is restricted by the classical nuclear import pathway. Myoblasts were transfected with inducible vectors for RFP-cNLS fusion proteins and differentiated for 2 d. (A) After differentiation, doxycycline was added to the culture medium with or without 5 µM importazole, an inhibitor of importin-β, and the distributions of RFP within individual Hoechst-stained myotubes were detected by epifluorescence microscopy. (B–D) The effect of importazole treatment on myonuclear propagation was quantified in myotubes expressing mCherry-cNLS (B), tdTomato-cNLS (C), and DsRed-cNLS (D). Distances and fluorescence intensities were normalized to the brightest myonucleus within each myotube. Data are binned by distance (bin size, 50 µm) and are represented as mean ± SEM (two-way ANOVA with Bonferroni posttest). The dotted line represents the approximate boundaries of the transfected myotubes. (Scale bars: 100 µm.) *P < 0.05; **P < 0.01; ***P < 0.001.
Myotube Atrophy and Hypertrophy Affect the Propagation of Nuclear Proteins.
Altered skeletal muscle homeostasis can lead to muscle atrophy, as a result of bed rest for example, or to muscle hypertrophy, as a result of weight training. However, it is unknown how altered muscle homeostasis alters the propagation of nuclear proteins. Muscle hypertrophy can be modeled in vitro using small-molecule activators of the mammalian target of rapamycin (mTOR) pathway. Specifically, the cation channel TRPV1 agonist capsaicin has been shown to activate mTOR by increasing intracellular calcium in a manner similar to load-induced calcium signaling (21, 22). Conversely, glucocorticoids have been shown to induce muscle atrophy in vitro. Dexamethasone is a corticosteroid that induces catabolism possibly by inhibition of insulin and insulin-like growth factor I, inhibition of protein synthesis machinery such as PI3K and mTOR, or induction of myostatin, cathepsin, calpain, and the ubiquitin-proteasome system (23). Therefore, capsaicin and dexamethasone were used to examine how perturbations in muscle size affect the propagation of nuclear proteins.
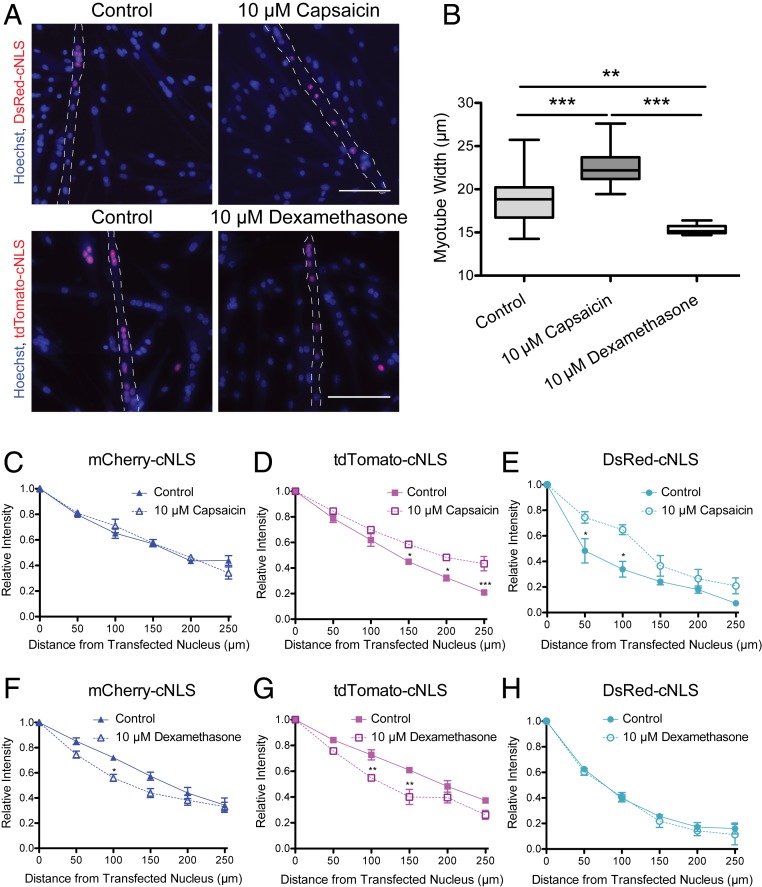
Myotubes were treated with either 10 µM capsaicin or 10 µM dexamethasone for 48 h to induce hypertrophy or atrophy, respectively, with doxycycline added during the final 24 h to induce expression of the RFP-cNLS fusion proteins. Capsaicin treatment increased average myotube width by 20% (from 18.9 µm to 22.7 µm) and altered the distribution of some RFP-cNLS fusion proteins (Fig. 4 A and B). Similar to importazole treatment, propagation of the mCherry-cNLS nuclear reporter was not affected by capsaicin-induced muscle hypertrophy (Fig. 4C); however, propagation of both the tdTomato-cNLS and DsRed-cNLS reporter proteins was extended significantly (Fig. 4 D and E). Dexamethasone treatment reduced average myotube width by 16% (from 18.9 µm to 15.8 µm) and also affected RFP-cNLS distribution. Both mCherry-cNLS and tdTomato-cNLS propagation was reduced by muscle atrophy (Fig. 4 F and G), while DsRed-cNLS distribution was not significantly altered (Fig. 4H). These data indicate that the effects of muscle morphology on the propagation of nuclear proteins may not be uniform, but instead may depend on the molecular weight of nuclear proteins.
Fig. 4.
Changes in myotube width affect the propagation of large RFP-cNLS fusion proteins. Myoblasts were transfected with inducible vectors for RFP-cNLS fusion proteins and differentiated for 2 d. Following myotube fusion, doxycycline was added to the culture medium to induce fusion protein expression. In some conditions, 10 µM capsaicin or 10 µM dexamethasone was included in the culture medium for the final 2 d of culture to induce muscle hypertrophy or atrophy. (A and B) The distributions of RFP-cNLS fusion proteins were imaged within Hoechst-stained myotubes (A), and myotube width was measured to quantify the effects of treatment (B). (C–E) The effect of capsaicin treatment on myonuclear propagation was quantified in myotubes expressing mCherry-cNLS (C), tdTomato-cNLS (D), and DsRed-cNLS (E). (F–H) The effect of dexamethasone treatment on myonuclear propagation was quantified in myotubes expressing mCherry-cNLS (F), tdTomato-cNLS (G), and DsRed-cNLS (H). Distances and fluorescence intensities were normalized to the brightest myonucleus within each myotube. Data are binned by distance (bin size, 50 µm) and are represented as mean ± SEM (two-way ANOVA with Bonferroni posttest). The dotted line represents the approximate boundaries of the transfected myotubes. (Scale bar: 100 µm.) *P < 0.05; **P < 0.01; ***P < 0.001.
Simulations of Nuclear Protein Transport Recapitulate In Vitro Experiments.
Intuitively, the propagation of nuclear proteins in a myotube is a competition between diffusive transport through the cytoplasm and facilitated nuclear import. If a protein diffuses sufficiently quickly or has a sufficiently low import rate, then it is able to “escape” the myonucleus that produced its transcript and enter other myonuclei. Conversely, a slow-moving protein that is rapidly imported might become localized to only the few nuclei around where it was produced. Yet it is not clear that these two processes are sufficient to explain the propagation of the RFP-cNLS constructs at all molecular weights. In fact, we have hypothesized that passive diffusion across the NPC may play an important role in explaining the broad propagation profile of the mCherry-cNLS construct, as well as its insensitivity to importazole.
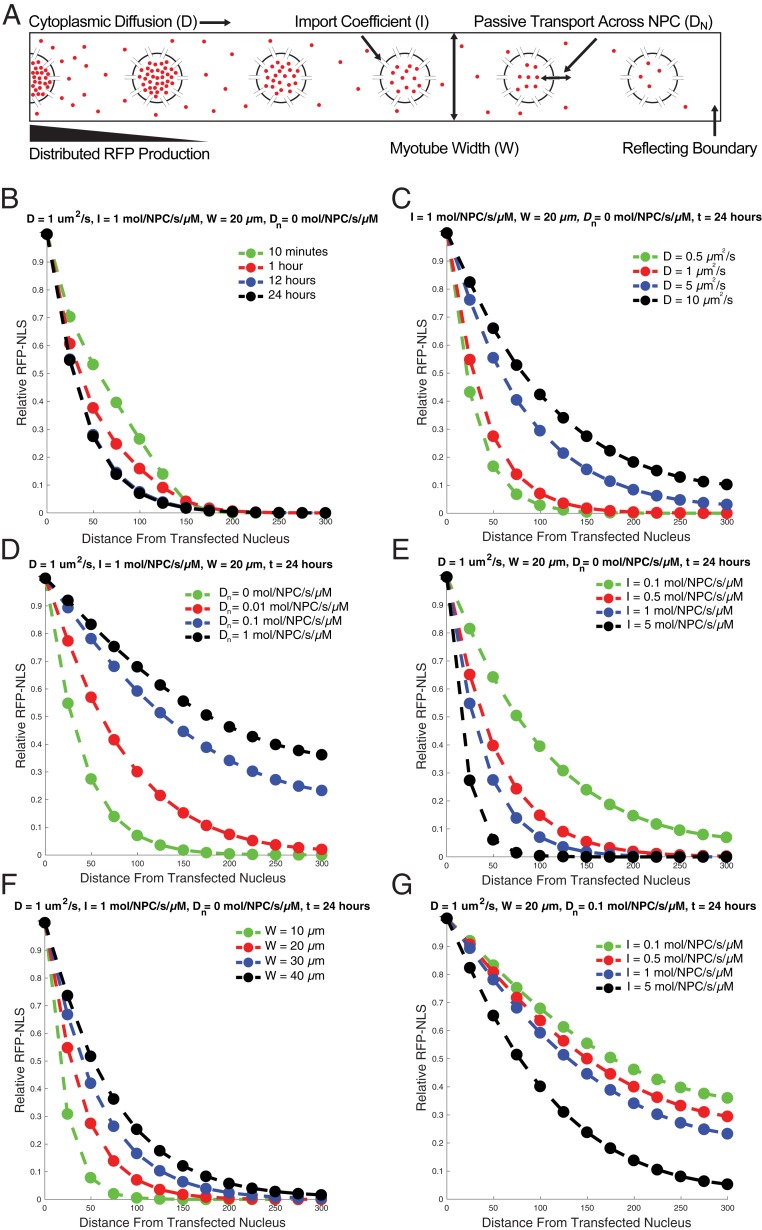
To address these questions, we built a mathematical model of RFP-cNLS transport inside a myotube (Fig. 5A) and used published values or empirical measurements to estimate initial parameters for the simulation (SI Appendix, Table S1 and Fig. S8). Since these parameters were approximations, we ran simulations exploring the parameter space to determine how these assumptions affected the behavior of the model. The simulation’s output was the relative concentration of RFP-cNLS in each myonucleus, allowing us to compare the results of the simulation with our experimental findings.
Fig. 5.
Simulations of RFP-cNLS diffusion and import recapitulate aspects of the in vitro data. (A) A mathematical model of RFP-cNLS transport was constructed that included cytoplasmic diffusion, facilitated nuclear import, diffusion across the NPC, and the ability to vary myotube width. The cytoplasmic production of RFP-cNLS decreased linearly from one boundary of the simulation (modeling a transfected nucleus at the center of the myotube), and a reflecting boundary was imposed at the terminus of the myotube. (B) The relative quantities of RFP-cNLS proteins within the simulated myonuclei were determined at different simulation times using a baseline parameter set (see also SI Appendix, Table S1). (C–G) The effects of varying different model parameters on RFP-cNLS propagation were determined for the cytoplasmic diffusion coefficient, D (C); the coefficient governing diffusion across the NPC, DN (D); the nuclear import coefficient, I (E); the myotube width, W (F); and the nuclear import coefficient with nonzero diffusion across the NPC (G).
Based on the initial parameter set, the mathematical model rapidly reached steady state (Fig. 5B). At early simulation time-points (10 min) the distribution of RFP-cNLS mirrors the distribution of the RFP transcript that was empirically measured (SI Appendix, Fig. S4) and which formed the basis for RFP production within the mathematical model. However, by 12 h, the simulation had reached steady state, leading to an exponential propagation profile similar to that observed using the DsRed-cNLS construct (Fig. 2B).
To model differences in RFP-cNLS molecular weight, we varied the cytoplasmic diffusion coefficient in the model. Increasing the diffusion coefficient from 0.5 µm2/s to 10 µm2/s increased the propagation distance of the simulated RFP-cNLS proteins (Fig. 5C). However, even at relatively large diffusion coefficients, the propagation curves were more exponential in shape rather than linear as had been observed experimentally with the smaller RFP-cNLS constructs (Fig. 2B). We had previously hypothesized that this difference might be caused by the ability of the smaller constructs to diffuse across the NPC, allowing them to “escape” the transfected nucleus; therefore, we tested allowing the simulated RFP-cNLS to diffuse across the NPC. At NPC diffusion coefficients >0.1 molecules/NPC/s/µM, the shape of the propagation curves became more linear, resembling the experimental data for the mCherry-cNLS and tdTomato-cNLS constructs (Fig. 5D). These data suggest that passive diffusion across the NPC plays an important role in regulating the propagation of nuclear proteins.
To model the in vitro experiments using importazole (Fig. 3), capsaicin, and dexamethasone (Fig. 4) treatments, we ran simulations that varied the facilitated import rate (Fig. 5E) and myotube width (Fig. 5F). Inhibiting the facilitated nuclear import and increasing the myotube width both extended the propagation of the simulated RFP-cNLS (Fig. 5 E and F), in agreement with our experimental findings for the large RFP-cNLS proteins. However, these data did not explain our observations using the mCherry-cNLS construct, which showed that this construct was insensitive to importazole and capsaicin treatment (Figs. 3B and 4C). Therefore, we examined how changes in the import rate affected the propagation of simulated RFP-cNLS proteins when the simulation included diffusion across the NPC (0.1 molecules/NPC/s/µM). Interestingly, under this condition, the propagation profiles were similar for import coefficients between 0.1 and 1 molecule/NPC/s/µM (Fig. 5G). These results imply that the ability for small proteins to diffuse across the NPC is sufficient to explain their relative insensitivity to importazole treatment. Taken together, these simulations suggest that the principle variables governing the propagation of the RFP-cNLS constructs were the cytoplasmic diffusion coefficient, the facilitated nuclear import rate, and the rate of passive diffusion across the NPC.
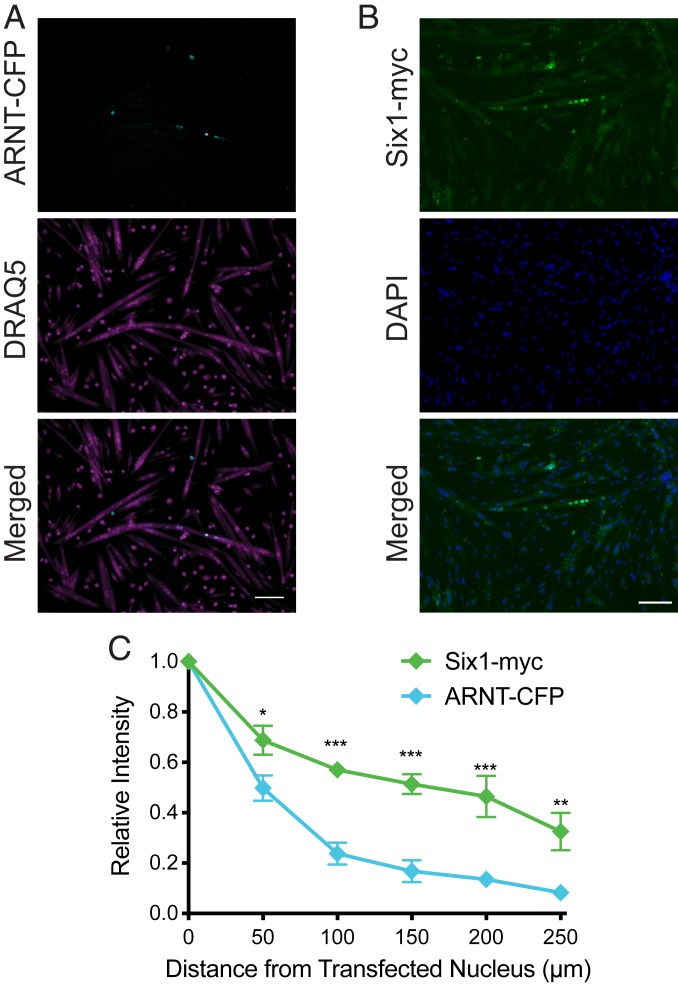
Propagation of ARNT and Six1 Is Similar to RFP-cNLS.
To determine whether skeletal muscle-relevant transcription factors can propagate in a manner similar to what had been predicted with the nuclear reporter proteins and the computational model, myoblasts were mosaically transfected with vectors for aryl hydrocarbon receptor nuclear translocator fused to cyan fluorescent protein (ARNT-CFP; ∼118 kDa) and sine oculis homeobox 1 protein fused to a myc tag (Six1-myc; ∼36 kDa). The ARNT transcription factor was chosen because it participates in hypoxic signaling, a relevant pathway in skeletal muscle, and contains a well-characterized NLS with similarities to the SV40 cNLS sequence (24). ARNT also efficiently localizes to the nucleus in a variety of contexts (24), suggesting that its NLS sequence is not independently regulated, similar to the RFP-cNLS reporter proteins. The Six1 transcription factor was chosen because of its role in muscle development, maintenance, and repair (25). It lacks a classical NLS but instead relies on residues within its homeodomain for nuclear localization (26). Both transcription factors possess protein and DNA-binding domains.
After 3 d, both ARNT-CFP and Six1-myc had propagated from the transfected myonucleus and entered neighboring myonuclei (Fig. 6 A and B). Compared with the RFP-cNLS reporter proteins, the propagation profiles of ARNT-CFP and Six1-myc most closely resembled those of the reporters most similar in molecular weight: DsRed-cNLS (110 kDa) and mCherry-cNLS (29 kDa), respectively (Fig. 6C). Specifically, myonuclei that were very close (0 to 50 µm) to the transfected nucleus contained only ∼45% of the ARNT-CFP of the transfected nucleus, similar to DsRed-cNLS and unlike the gradual propagation profiles of the smaller reporter proteins and Six1-myc. These data suggest that our results with simple nuclear reporter proteins can help predict the propagation profiles of skeletal muscle-relevant transcription factors.
Fig. 6.
Propagation of ARNT-CFP and Six1-myc is similar to that of RFP-cNLS fusion proteins. Myoblasts were transfected with an expression vector for either aryl hydrocarbon receptor nuclear translocator fused to cyan fluorescent protein (ARNT-CFP; ∼118 kDa) or sine oculis homeobox 1 bearing a myc-tag (Six1-myc). (A and B) After 3 d of differentiation, the distributions of ARNT-CFP within DRAQ5-stained myotubes (A) or Six1-myc within DAPI-stained myotubes (B) were visualized by epifluorescence microscopy. (C) Transcription factor propagation was quantified by measuring the positions and average intensity of myonuclei within individual transfected myotubes. Distances and fluorescence intensities were normalized to the brightest myonucleus within each myotube (assumed to be the transfected nucleus). Data are binned by distance (bin size, 50 µm) and are represented as mean ± SEM (two-way ANOVA with Bonferroni posttest). (Scale bar: 100 µm.) *P < 0.05; **P < 0.01; ***P < 0.001.
Discussion
In this study, we have introduced the concept of “propagation” to describe the distance proteins travel within large, multinucleated cells and specifically examined how nuclear proteins become distributed among myonuclei within myotubes. Using three closely related RFP-cNLS fusion proteins, we determined that molecular weight affects the propagation of nuclear proteins in myotubes. Surprisingly, there were only modest differences in the propagation of the two smaller fluorescent reporters (28.8 and 55 kDa), both of which decreased steadily from the transfected myonucleus. By contrast, the largest reporter (110 kDa) exhibited a large initial decrease in signal before beginning a gradual decline (Fig. 2). We hypothesized that this biphasic behavior may be a result of its inability to diffuse across the NPC, which is limited in proteins larger than 60 kDa (17). Since the largest reporter likely cannot diffuse across the NPC, it may become trapped within the first myonucleus it enters. By contrast, the smaller nuclear reporters may diffuse back into the cytoplasm and rejoin the pool of diffusing proteins moving in the cytoplasm. This observation likely has important consequences in muscle, especially considering the range of transcription factor molecular weights. Using the Human Protein Atlas, we analyzed the predicted molecular weights of 70 transcription factors that are highly expressed in human skeletal muscle. The 25th and 75th percentiles of the molecular weights were 50.75 kDa and 83.5 kDa, respectively, although the range extended from 13 kDa to 283 kDa (median, 62.5 kDa). Gene Ontology analysis of the 32 largest (66 to 283 kDa) and smallest (13 to 58 kDa) transcription factors found that the smaller cohort was significantly enriched in factors that regulate cellular response to soluble stimuli (11 genes total; P = 2.69e-4), including hormones (six genes), oxygen-containing compounds (eight genes), acids (four genes), and metal ions (four genes). The larger cohort was significantly enriched in transcription factors that regulate DNA organization, including covalent chromatin modifications (seven genes; P = 1.39e-6). It is possible that this segregation in functionality allows myonuclei to use small transcription factors to coordinate a cell-wide response to systemic signals (soluble compounds) while maintaining transcriptional autonomy through localized reorganization of DNA.
In our study, the propagation of larger nuclear proteins was strongly affected by their nuclear import rate. Using importazole, a small-molecule inhibitor of the classical nuclear import pathway (20), we found that RFP-cNLS fusion proteins larger than 55 kDa propagated significantly farther when nuclear import was slowed (Fig. 3). Inhibition of nuclear import gives nuclear proteins a longer lifetime in the cytoplasm, where they can diffuse down the length of the myotube before becoming sequestered into a myonucleus. Interestingly, nuclear import is slowed in myofibers following injury (18), which may increase the propagation distances of all large nuclear proteins. Perhaps slowing nuclear import allows nuclear proteins that originate from uninjured portions of the myofiber to more easily reach myonuclei at the site of injury and participate in regeneration.
The import rate of nuclear proteins may vary based on differences in NLS and posttranslational modifications that regulate accessibility of the NLS. In fact, the effective import rate of a fluorescent reporter fused with different NLS peptides can vary by >10-fold based on the specific NLS (27). Moreover, unlike molecular weight, the nuclear import rate may provide a simple evolutionary mechanism for tuning the propagation distances of nuclear proteins without affecting other functions (e.g., DNA binding, enzymatic activity). In fact, tuning NLS peptides may prove useful in designing gene therapies in skeletal muscle; for example, induced expression of exercise-inducible nuclear proteins PGC-1α/β and JUNB has been proposed to treat muscle-wasting diseases (28). However, these treatments may be limited by the relatively small number of myonuclei that are infected, leading to a locally confined treatment. Our results suggest that this limitation might be overcome by modifying the NLS peptides of the induced nuclear proteins to slow the rate of nuclear import and extend their propagation. This could allow a relatively small number of myonuclei, distributed over the length of the muscle, to affect the behavior of the entire muscle fiber.
Using capsaicin to induce myotube thickening in vitro, we found that muscle hypertrophy extends the propagation of large nuclear proteins (Fig. 4). Since there were no undifferentiated progenitor cells in our experiments, our in vitro results reflect hypertrophy without the fusion of additional myonuclei. However, during muscle hypertrophy in vivo, myofibers first increase the cross-sectional area and then activate resident satellite cells to increase myonuclear number (29). In fact, it has been hypothesized that the initial increase in cytoplasm per myonucleus (myonuclear domain) during hypertrophy produces a signal for new myonuclei to fuse (29). Indeed, our mathematical model showed that increasing myonuclear density could offset the “dilution” effects of muscle hypertrophy and restore the original propagation profile of nuclear proteins (SI Appendix, Fig. S9). Taken together, our data suggest that nuclear protein propagation in myotubes can be a signal that alerts myonuclei to changes in cell morphology and dissipates once the homeostatic myonuclear domain is restored.
Most transcription factors expressed in skeletal muscle possess binding domains for proteins and DNA, while RFPs are relatively inert with respect to the intracellular constituents with which they can interact. While intermolecular association could conceivably have significant effects on nuclear protein propagation, our results with ARNT and Six1 suggest that the predominant determinants of transcription factor propagation in muscle are the cytoplasmic diffusion coefficient, the facilitated nuclear import rate, and the rate of passive diffusion across the NPC. However, nuclear receptors (30) and other ligand-inducible transcription factors that require cofactors for nuclear translocation, as well as proteins that exploit active cytoplasmic transport mechanisms (31), would likely exhibit propagation profiles that deviate from those presented here.
In summary, we have used a mosaic transfection model to visualize the propagation of nuclear proteins within myotubes. Taken together, this work provides a basis for making de novo predictions of nuclear protein propagation within myotubes. This information may help explain how muscle cells coordinate gene expression among many myonuclei and provide some important design parameters (e.g., infection rate, spatial distribution) for engineering gene therapies in skeletal muscle.
Materials and Methods
Cell Culture.
Primary mouse myoblasts were isolated as described previously (32) and cultured at 37 °C with 5% CO2 in a humidified incubator. Myoblasts were maintained in growth medium composed of 20% (vol/vol) FBS (Thermo Fisher Scientific, 10270), 1% (vol/vol) penicillin-streptomycin (Thermo Fisher Scientific, 15140122), and 2.5 ng/mL basic fibroblast growth factor (Thermo Fisher Scientific, 13256029) in 1:1 DMEM (Thermo Fisher Scientific, 41965):Ham’s F-10 Nutrient Mix (Thermo Fisher Scientific, 22390) on plates coated with 50 µg/mL type I collagen (Corning, 354249).
To induce myotube fusion, myoblasts were seeded at a density of 1.7e5 cells/cm2 in growth medium onto well plates coated with 0.1 mg/mL poly-l-lysine hydrobromide (Sigma-Aldrich, P6282) and a solution of 50 µg/mL type I collagen, 10 µg/mL fibronectin (Sigma-Aldrich, F1141), and 10 µg/mL laminin (Sigma-Aldrich, L2020) in PBS (Thermo Fisher Scientific, 14190). Once the myoblasts had attached (∼1 h), the medium was replaced with Opti-MEM (Thermo Fisher Scientific, 51985034), and the cells were transfected with 150 ng of the indicated plasmid using DNA/reducible poly(amido amine) nanoparticles, as described previously (33).
To induce expression of the RFP-cNLS expression vectors, 2 µg/mL doxycycline (Sigma-Aldrich, D9891-1G) was added to differentiation medium for 24 h, as indicated. When indicated, 5 µM importazole (Sigma-Aldrich, SML0341) was added to differentiation medium for 24 h to partially inhibit the classical nuclear import pathway. To induce myotube hypertrophy, 10 µM capsaicin (Sigma-Aldrich, 12084) was added to differentiation medium for 48 h. To induce myotube atrophy, 10 µM dexamethasone (Sigma-Aldrich, D4902) was added to the differentiation medium for 48 h.
Supplementary Material
Acknowledgments
This work was supported by a Human Frontier Science Program Young Investigator Award (RGY0082-2014, to J.L.R. and A.I.T.), the Whitaker International Program (H.T.-W. and C.L.G.), and the Swedish Research Council (Grants 2015-03520, to A.I.T.; 2016-0785, to J.L.R.; and 2016-04478, to M.M.S.). Cell imaging was performed at the Biomedicum Imaging Core, and cell sorting was performed at the Biomedicum Flow Cytometry Core Facility with support from Karolinska Institutet.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: All plasmids generated for this work are available through the Addgene repository (ID codes 138527–138533). Data are available from the Swedish National Data Service (https://doi.org/10.5878/2hjy-e586).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1919600117/-/DCSupplemental.
References
- 1.Newlands S., et al. , Transcription occurs in pulses in muscle fibers. Genes Dev. 12, 2748–2758 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen D. L., Roy R. R., Edgerton V. R., Myonuclear domains in muscle adaptation and disease. Muscle Nerve 22, 1350–1360 (1999). [DOI] [PubMed] [Google Scholar]
- 3.Price F. D., Kuroda K., Rudnicki M. A., Stem cell-based therapies to treat muscular dystrophy. Biochim. Biophys. Acta 1772, 272–283 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Briguet A., Ruegg M. A., The Ets transcription factor GABP is required for postsynaptic differentiation in vivo. J. Neurosci. 20, 5989–5996 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fontaine B., Sassoon D., Buckingham M., Changeux J. P., Detection of the nicotinic acetylcholine receptor alpha-subunit mRNA by in situ hybridization at neuromuscular junctions of 15-day-old chick striated muscles. EMBO J. 7, 603–609 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brenner H. R., Witzemann V., Sakmann B., Imprinting of acetylcholine receptor messenger RNA accumulation in mammalian neuromuscular synapses. Nature 344, 544–547 (1990). [DOI] [PubMed] [Google Scholar]
- 7.O’Connor R. S., Mills S. T., Jones K. A., Ho S. N., Pavlath G. K., A combinatorial role for NFAT5 in both myoblast migration and differentiation during skeletal muscle myogenesis. J. Cell Sci. 120, 149–159 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Abbott K. L., Friday B. B., Thaloor D., Murphy T. J., Pavlath G. K., Activation and cellular localization of the cyclosporine A-sensitive transcription factor NF-AT in skeletal muscle cells. Mol. Biol. Cell 9, 2905–2916 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferri P., et al. , Expression and subcellular localization of myogenic regulatory factors during the differentiation of skeletal muscle C2C12 myoblasts. J. Cell. Biochem. 108, 1302–1317 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Artaza J. N., et al. , Endogenous expression and localization of myostatin and its relation to myosin heavy chain distribution in C2C12 skeletal muscle cells. J. Cell. Physiol. 190, 170–179 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Hall M. N., Corbett A. H., Pavlath G. K., Regulation of nucleocytoplasmic transport in skeletal muscle. Curr. Top. Dev. Biol. 96, 273–302 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalderon D., Richardson W. D., Markham A. F., Smith A. E., Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature 311, 33–38 (1984). [DOI] [PubMed] [Google Scholar]
- 13.Shaner N. C., et al. , Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22, 1567–1572 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Strack R. L., et al. , A noncytotoxic DsRed variant for whole-cell labeling. Nat. Methods 5, 955–957 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eisner V., Csordás G., Hajnóczky G., Interactions between sarco-endoplasmic reticulum and mitochondria in cardiac and skeletal muscle—Pivotal roles in Ca2+ and reactive oxygen species signaling. J. Cell Sci. 126, 2965–2978 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan Y., Simmen T., Mechanistic connections between endoplasmic reticulum (ER) redox control and mitochondrial metabolism. Cells 8, E1071 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang R., Brattain M. G., The maximal size of protein to diffuse through the nuclear pore is larger than 60 kDa. FEBS Lett. 581, 3164–3170 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cutler A. A., Jackson J. B., Corbett A. H., Pavlath G. K., Non-equivalence of nuclear import among nuclei in multinucleated skeletal muscle cells. J. Cell Sci. 131, jcs207670 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall M. N., Griffin C. A., Simionescu A., Corbett A. H., Pavlath G. K., Distinct roles for classical nuclear import receptors in the growth of multinucleated muscle cells. Dev. Biol. 357, 248–258 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soderholm J. F., et al. , Importazole, a small molecule inhibitor of the transport receptor importin-β. ACS Chem. Biol. 6, 700–708 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito N., Ruegg U. T., Kudo A., Miyagoe-Suzuki Y., Takeda S., Activation of calcium signaling through Trpv1 by nNOS and peroxynitrite as a key trigger of skeletal muscle hypertrophy. Nat. Med. 19, 101–106 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Ito N., Ruegg U. T., Kudo A., Miyagoe-Suzuki Y., Takeda S., Capsaicin mimics mechanical load-induced intracellular signaling events: Involvement of TRPV1-mediated calcium signaling in induction of skeletal muscle hypertrophy. Channels (Austin) 7, 221–224 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han D. S., Yang W. S., Kao T. W., Dexamethasone treatment at the myoblast stage enhanced C2C12 myocyte differentiation. Int. J. Med. Sci. 14, 434–443 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eguchi H., Ikuta T., Tachibana T., Yoneda Y., Kawajiri K., A nuclear localization signal of human aryl hydrocarbon receptor nuclear translocator/hypoxia-inducible factor 1beta is a novel bipartite type recognized by the two components of nuclear pore-targeting complex. J. Biol. Chem. 272, 17640–17647 (1997). [DOI] [PubMed] [Google Scholar]
- 25.Le Grand F., et al. , Six1 regulates stem cell repair potential and self-renewal during skeletal muscle regeneration. J. Cell Biol. 198, 815–832 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu W., et al. , Subcellular localization of different regions of porcine Six1 gene and its expression analysis in C2C12 myoblasts. Mol. Biol. Rep. 39, 9995–10002 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Timney B. L., et al. , Simple kinetic relationships and nonspecific competition govern nuclear import rates in vivo. J. Cell Biol. 175, 579–593 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cohen S., Nathan J. A., Goldberg A. L., Muscle wasting in disease: Molecular mechanisms and promising therapies. Nat. Rev. Drug Discov. 14, 58–74 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Petrella J. K., Kim J. S., Mayhew D. L., Cross J. M., Bamman M. M., Potent myofiber hypertrophy during resistance training in humans is associated with satellite cell-mediated myonuclear addition: A cluster analysis. J. Appl. Physiol. 104, 1736–1742 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Kupr B., Schnyder S., Handschin C., Role of nuclear receptors in exercise-induced muscle adaptations. Cold Spring Harb. Perspect. Med. 7, a029835 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carbonaro M., Escuin D., O’Brate A., Thadani-Mulero M., Giannakakou P., Microtubules regulate hypoxia-inducible factor-1α protein trafficking and activity: Implications for taxane therapy. J. Biol. Chem. 287, 11859–11869 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruas J. L., et al. , A PGC-1α isoform induced by resistance training regulates skeletal muscle hypertrophy. Cell 151, 1319–1331 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin C., et al. , Novel bioreducible poly(amido amine)s for highly efficient gene delivery. Bioconjug. Chem. 18, 138–145 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.