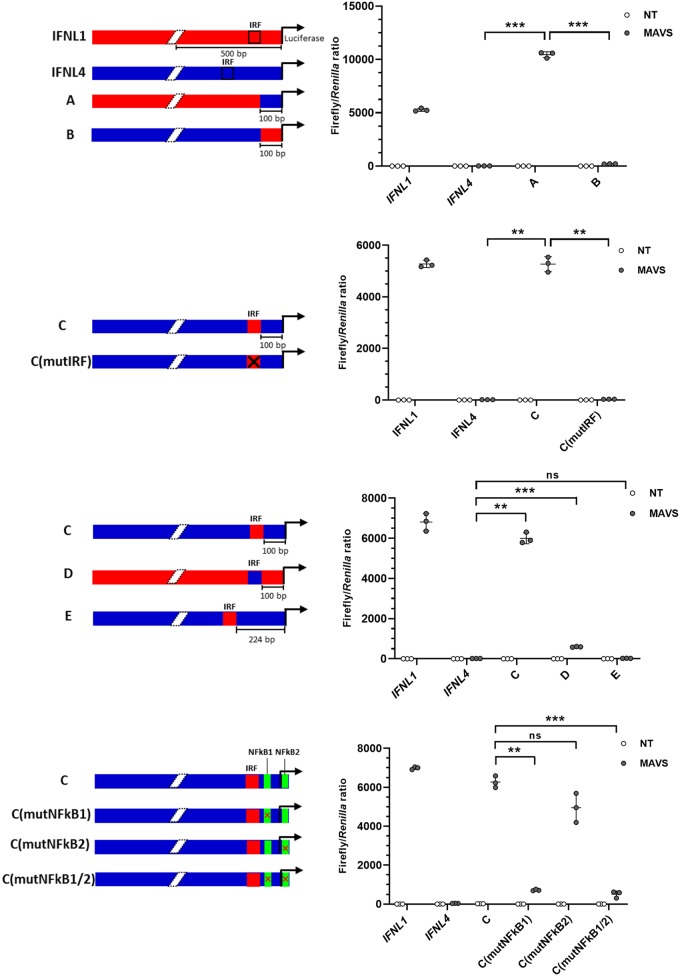
FIG 6.
Insertion of a functional IRF binding site in the IFNL4 promoter renders it virus inducible. (A to D) HEK293T cells were cotransfected with a pGL3.1 plasmid containing the IFNL1 or IFNL4 promoter or chimeras thereof, a plasmid constitutively expressing the Renilla luciferase gene, and a plasmid constitutively expressing MAVS. At 24 h posttransfection, firefly/Renilla luciferase activity was quantified. One representative out of two independent experiments is shown, each with biological triplicates. The data are presented as a scatter plot with mean ± SD (n = 3). (A) In chimeras A and B, the first 100 bp upstream of the TSS has been swapped between the IFNL1 and IFNL4 promoters, as indicated by the colors in the diagram. (B) In chimera C, the IRF binding site from IFNL1 was inserted into the IFNL4 promoter at the same relative position it occupied in IFNL1. In chimera C (mutIRF), the IFNL1 IRF binding site in chimera C was mutated to render it inactive. (C) In chimera D, the IRF binding site in IFNL1 was replaced with the putative IRF binding site from IFNL4. In chimera E, the putative IRF binding site in IFNL4 was replaced with the IRF binding site from IFNL1. (D) In chimera C (mutNF-κB1), the NF-κB binding site located upstream of the TSS in IFNL4 was mutated to render it inactive. In chimera C (mutNF-κB2), the NF-κB binding site located downstream of the TSS in IFNL4 was mutated to render it inactive. In chimera C (mutNF-κB1/2) both NF-κB binding sites were simultaneous mutated. Statistical significance was determined using analysis of variance (ANOVA) and Dunnett’s T3 multiple-comparison test. ***, 0.0001 < P < 0.001; **, 0.001 < P < 0.01; ns, P ≥ 0.05.