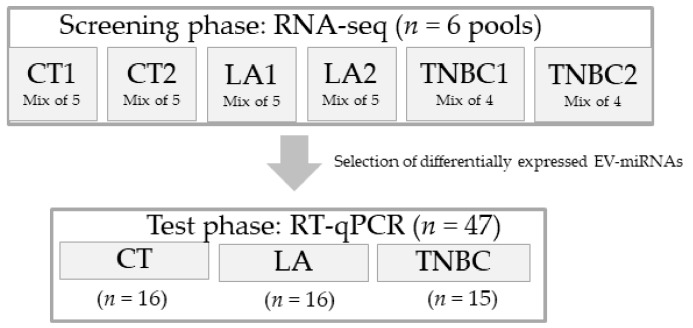
Figure 1.
Schematic diagram of the workflow of this study. This study was designed in two phases. During the screening phase we performed RNA sequencing (RNA-seq) analysis in six pools of sample composed of two sample per group (Control (CT), Luminal A (LA), and Triple-negative (TNBC). During the test phase, a selection of EV-miRNAs found to be differentially expressed in the RNA-seq analysis was chosen to be tested in 47 individual patient samples’ by quantitative real-time PCR (RT-qPCR).