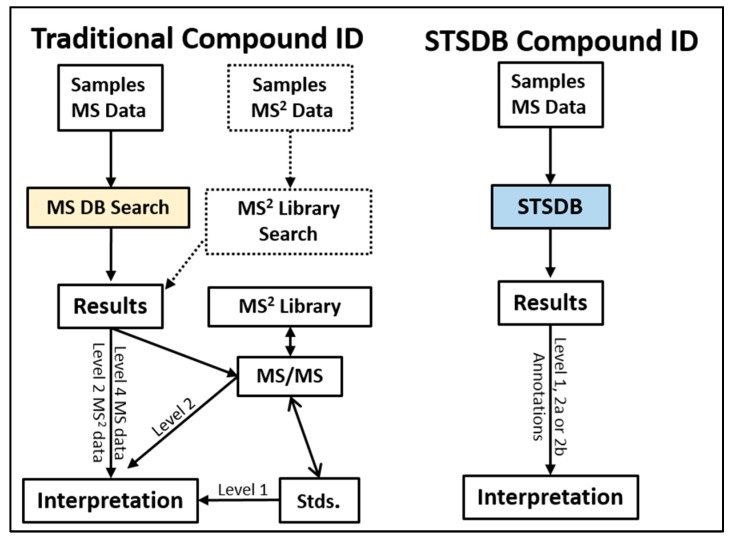
Figure 1.
Traditional database (DB) vs. proposed sample type specific database (STSDB) searching workflows. The traditional workflow for annotating metabolomics data using databases is shown on the left. Typically, an MS database or MS/MS library search is conducted for initial annotation of compounds. Results from MS and MS/MS searches may be combined to aid in interpretation and authentic standards are used to confirm the identity of the compound. As detailed below, the confidence with which a compound is assigned a name (i.e., annotated) is currently assigned to 4 levels ranging from “unknown” (Level 4) to “Identified” (Level 1). Level 1 confidence currently requires matching to a standard (Std). Compound identification using the sample type specific database (STSDB) approach is shown on the right whereby MS data (i.e., mass, isotope ratios, and formulas) is used to search an STSDB. STSDBs store information on compounds such that higher levels of confidence are obtained following a single search.