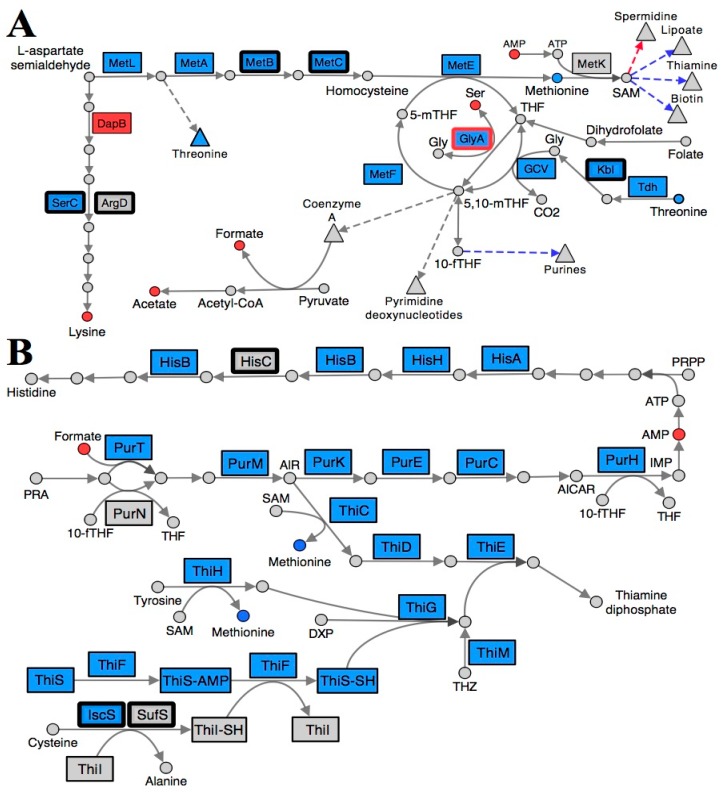
Figure 2.
Folate and S-adenosylmethionine pathways are reorganized in a S. enterica ridA mutant. Metabolites (circles) and gene products (rectangles) involved in (A) lysine, methionine, and folate biosynthesis and (B) purine, histidine, and thiamine biosynthesis are depicted, with each arrow representing a biochemical step in the pathway. Blue shading denotes a significantly (FDR < 0.05) increased abundance of the respective metabolite/transcript in ridA cells, while red shading indicates a significantly decreased abundance in ridA cells. Grey gene products showed no significant difference between wild-type and ridA cells, while grey metabolites were either not identified or not significantly altered. A bold border around a gene product designates enzymes that use PLP as a cofactor. GlyA is a known target for 2AA damage and is shown with a red border. Dashed arrows and triangles indicated general pathways which utilize the metabolite at the base of the arrow, while blue and red colored arrows denote genes involved in biosynthesis of that compound up- or down-regulated, respectively.