Abstract
Verticillium wilt, caused by the ascomycete fungus Verticillium dahliae (Vd), is a devastating disease of numerous plant species. However, the pathogenicity/virulence-related genes in this fungus, which may be potential targets for improving plant resistance, remain poorly elucidated. For the study of these genes in Vd, we used a well-established host-induced gene silencing (HIGS) approach and identified 16 candidate genes, including a putative adenylate kinase gene (VdAK). Transiently VdAK-silenced plants developed milder wilt symptoms than control plants did. VdAK-knockout mutants were more sensitive to abiotic stresses and had reduced germination and virulence on host plants. Transgenic Nicotiana benthamiana and Arabidopsis thaliana plants that overexpressed VdAK dsRNAs had improved Vd resistance than the wild-type. RT-qPCR results showed that VdAK was also crucial for energy metabolism. Importantly, in an analysis of total small RNAs from Vd strains isolated from the transgenic plants, a small interfering RNA (siRNA) targeting VdAK was identified in transgenic N. benthamiana. Our results demonstrate that HIGS is a promising strategy for efficiently screening pathogenicity/virulence-related genes of Vd and that VdAK is a potential target to control this fungus.
Keywords: host-induced gene silencing (HIGS), Verticillium dahliae, pathogenicity factor, adenylate kinase
1. Introduction
Verticillium dahliae (Vd), the causal agent of Verticillium wilt (Vw), is a destructive fungal pathogen infecting over 400 plant species, including important ornamental, horticultural, agronomical, and woody plants [1]. Symptoms are not uniform among different plant species [2], and in general, the fungus is difficult to control because it survives in the plant vascular system and infects the hosts via infested soil and diseased plant debris [3]. Once the plants are infected, no fungicides can cure the plants [4]. Vd can cause severe economic losses worldwide. In China, approximately 250–310 million US dollar losses have been reported for cotton annually due to Vd [5]. Considerable studies on Vw and the associated fungi and the public release of the genomic sequence of Vd and its sister fungus V. alfalfae, have enabled the identification of candidate pathogenicity and virulence genes [6,7,8]. However, the complexity of the pathogenic mechanism is a serious constraint on developing effective management strategies. In addition, we still have little knowledge about virulence factors in Verticillium species [9,10,11,12], although a few genes from Vd are known to be involved in its virulence of this fungus. For instance, the deletion of the β-1,6-endoglucanase gene substantially reduced disease symptoms in eggplant by compromising the utilization of cellulose [13]. The adhesion protein Vta2 and transmembrane protein mucin Msb, which serve as positive regulators, enable microsclerotial formation, adhesive capacity, invasive growth, and/or sporulation [10,14]. VdMcm1, a MADS-box transcription factor, has multiple biological functions during spore and microsclerotial production, secondary metabolism, and virulence [15]. As a member of the Myosin V family, VdMyo5 may participate in vesicle transport to regulate vegetative growth and is essential for full virulence [16]. Although multiple genes playing a role in signal pathways, development, and nutrition have been reported [17,18,19,20], few have been confirmed as feasible candidates for the control of Vd in plants.
A host-induced gene silencing (HIGS) method for in planta RNAi silencing of a parasitic fungal gene achieved a breakthrough as a pathogen-derived resistance strategy to control fungi and other microorganisms [21,22,23,24,25]. With this method, manipulation of the accumulation of dsRNA that targets fungal transcripts in barley (Hordeum vulgare) and wheat (Triticum aestivum) inhibited the development of the powdery mildew fungus Blumeria graminis [26]. HIGS, guided by Barley stripe mosaic virus (BSMV), interfered with the expression of three predicted pathogenicity genes, thus suppressing invasion and colonization by the wheat leaf rust fungus, Puccinia triticina [27]. Synthetic dsRNAs could compromise expression of the targeted fungal gene and inhibited conidial germination of Fusarium oxysporum f. sp. cubense and Mycosphaerella fijiensis [28]. In a transgenic banana that expressed a hairpin RNA against velvet and Fusarium transcription factor 1 genes of F. oxysporum f. sp. cubense (Foc), resistance was improved for 8 months, indicating potential use for breeding [29]. HIGS-assisted silencing of β-1,3-glucan synthase gene FcGls1 caused elevated resistance against Fusarium head blight [30]. In recent independent studies, the HIGS strategy was deployed to interfere with desirable target genes of Vd, and new virulence factors, e.g., RGS1, Ave1, Sge1, and NLP1, were identified [31,32]. For instance, HIGS-assisted interference with Vd RGS1 conferred elevated resistance in cotton [31].
In previous studies, we generated transgenic plants by overexpressing dsRNA against VdAAC (ADP, ATP carrier: Responsible for transferring ATP from the mitochondria into the cytoplasm) and VdSTT3 (oligosaccharyl transferase subunit, playing an essential role in glycoprotein modification) to improve resistance against Vd [33,34]. To explore more candidate genes from Vd to target and further verify the use of the HIGS system, here we (1) screened for Vd candidate pathogenicity/virulence factors to take advantage of genomics data and the well-established HIGS approach; (2) characterized adenylate kinase (VdAK) functions in virulence; (3) evaluated the resistance level of transgenic Nicotiana benthamiana, and Arabidopsis thaliana harboring dsVdAK against Vd and (4) demonstrated that small interfering RNAs (siRNAs) can enter fungi from within the plants and improve the resistance of host plants.
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
N. benthamiana plants were cultivated in pots containing sterilized soil (peat compost: vermiculite, 1:1, w/w) in a greenhouse (25 ± 2 °C and 75% relative humidity, 16 h light and 8 h dark). A. thaliana ecotype Columbia-0 was sown and grown in the growth chamber (23 ± 2 °C, 70% relative humidity, 16 h light and 8 h dark, light intensity 4000 lx).
2.2. Construction of HIGS and Transforming Plasmids and Plant Transformation
Total RNA was isolated using an RNA extraction kit (YPHBio, Tianjin, China), and 1 μg of the total RNA was used to synthesize the first-strand cDNA using a reverse transcription kit (TransGen, Beijing, China) according to the manufacturer’s instructions. The construction of HIGS plasmids was initiated using PCR amplification of target fragments of 400 to 600 bp. These sequence-verified target fragments were cloned into tobacco rattle virus (TRV) vector via double-digestion with BamHI and EcoRI and ligation with T4 ligase using standard protocols. TRV:00 or its derivatives were independently transformed into Agrobacterium tumefaciens strain GV3101 by electroporation [35]. Subsequently, the constructs were agroinfiltrated into seedlings with a needleless syringe [36,37]. The primers used for PCR amplification are listed in Supplementary Table S1.
Gateway cloning technology was used to construct the plasmid for expressing dsRNA of VdAK. The target fragment was produced by PCR amplification from Vd cDNA, then cloned into the intermediate vector pDONR207. The expression cassette pK7GWIWG2(I)::VdAK was obtained via attL and attR (LR) recombination reaction between the entry plasmid pDONR207::VdAK and the destination vector pK7GWIWG2(I) using Gateway LR Clonase II Enzyme Mix (Invitrogen, San Diego, CA). The sequence-verified plasmid was transformed into A. tumefaciens strain LBA4404 by electroporation to transform N. benthamiana and A. thaliana plants using a leaf disc co-cultivation method and floral dip method, respectively [38,39,40]. The primers are listed in Supplementary Table S2.
2.3. Assessment of Disease Resistance
The highly virulent wild-type Vd strain V991 was shake-cultured in a complete medium (CM) containing 50 mg/L ampicillin and 50 mg/L kanamycin at 25 °C for 6 days. The roots of seedlings were incubated for 2 min in a suspension of 1 × 106 spores/mL, and immediately replanted into new pots. Disease severity (DS) at 10 to 12 days post-inoculation (dpi) was scored using previously described methods [33,41] as 0, no wilt; 1, less than 2 leaves wilting; 2, 3–5 leaves wilting; 3, more than 5 leaves wilting or chlorotic; and 4, plant death or near death, to calculate a disease index (DI): DI = [∑ (No. of plants × DS score)/(Total no. of plants × Highest DS score)] ×100.
The fungal biomass in tissues was quantified using a slight modification of a previously described method [12]. In brief, DNA was extracted using the Plant Genomic DNA Kit (TIANGEN, Beijing, China) separately from infected roots, stems (0–3 cm above the ground surface), and leaves of 5 plants. ITS1 and ITS2 regions of the ribosomal RNA genes (Z29511) of Vd were amplified to quantify fungal DNA as previously described [12].
Hyphal development in roots of host plants after inoculation with Vd-GFP (green fluorescent protein) and ΔVdAK-GFP strains (described next) was assessed at 5 dpi using a confocal microscope (Zeiss LSM 700, Jena, Germany).
2.4. VdAK Gene Disruption, Complementation, and VdAK-GFP Mutant Strains
The VdAK gene knockout plasmid was generated using a previously described method [33]. Briefly, ~1.2-kb upstream and downstream flanking fragments of VdAK and hygromycin resistance (HPT) gene expression cassette were fused by overlap PCR. The knockout fragment was obtained using nested PCR by fusion PCR product as the template, then cloned into the pGKO2 vector by attB and attP (BP) recombination reaction using Gateway BP Clonase II Enzyme Mix (Invitrogen, San Diego, CA, USA).
For the construction of the ΔVdAK-GFP strains, the neomycin resistance (NeoR) gene expression cassette amplified from pCAM-neo with TrpC promoter and TrpC terminator was inserted into the expression plasmid pCAMBIA1302 via XbaI and BstEII restriction sites. The GFP expression cassette was cloned into the plasmid via the XbaI and KpnI restriction sites to generate pCAMBIA1302::neo::GFP. Then, the GFP open reading frame (ORF) was replaced with the VdAK ORF via the ScaI and PstI restriction enzyme sites to generate pCAMBIA1302::neo::VdAK for ΔVdAK-C.
Transformants, including the ΔVdAK, ΔVdAK-C strains and ΔVdAK-GFP, were obtained by protoplast transformation [42]. Positive transformants were selected by RT-PCR and parallel antibiotic resistance. The primers used in these constructions are listed in Table S3.
2.5. Stress Treatments
A 10 µL drop of 1 × 106 spores/mL of the respective strains was placed on a plate of Czapek-Dox agar amended with 0.5 M NaCl or 0.5 M sorbitol or treated with a 10 s pulse of 302 nm UV light [43]. Colony diameters were measured after 4 weeks. For estimating spore production, 3 mL of sterilized water was added to each plate, which was gently shaken to release the spores [12]. The spores were counted using a hemacytometer and light microscope (OLYMPUS BX52, Tokyo, Japan).
2.6. RT-qPCR
The expression of target genes was quantified by RT-qPCR using a 7500 Real Time PCR System (Applied Biosystems, Foster City, CA, USA) and TransStart Top Green qPCR SuperMix (TransGen, Beijing, China) according to the manufacturer’s instructions. The 2-∆∆Ct method was used for relative quantification of transcripts for each gene [44]; the N. benthamiana housekeeping gene (Nbactin), A. thaliana housekeeping gene (AtEF-1α) or Vd housekeeping gene (Vdactin) was used as an internal control. The primers are listed in Supplementary Table S4.
2.7. Small RNA (sRNA) Sequencing and Data Analysis
The stems of Vd-infected seedlings were surface-disinfested and placed on potato dextrose agar (PDA). After 1 week, any fungal colony that had grown from the infected stems was cultured on PDA for observation and in CM broth for RNA extraction. sRNA library construction and sequencing were performed by Novogene (http://www.novogene.com/). Total sRNAs were mapped to the published genomes of Verticillium (Broad Institute, https://www.broadinstitute.org/). The targeting relationship between reads and targeted sequences of VdAK was predicted by miRanda (http://www.microrna.org/microrna/home.do). Information on sRNAs from Vd isolated from Vd-infected seedlings is listed in Table S5.
2.8. Statistical Analysis
Data from 3 independent experiments were analyzed using Duncan’s multiple range test using SPSS Statistics 17.0 software (SPSS, Chicago, IL, USA).
3. Results
3.1. HIGS Candidate Pathogenicity Factors Were Selected Based on Available Protein-Encoding Genes
To identify the pathogenicity factor genes that are required for Vd virulence, we employed a well-established virus-guided HIGS system [45]. For the classification and annotation of the 10,535 publicly available predicted protein-encoding genes of the Vd strain VdLs.17 [6], GO analysis was carried out (Figure S1). Based on SwissProt and Blast results, these genes were divided into cellular components, molecular functions, and biological processes; 92 fungal genes involved in diverse biological processes (energy, metabolism, development, secreted protein, and others) were considered as candidates for HIGS in N. benthamiana system (Table S1). They had no sequence similarity in an analysis of sequence similarity between these genes and the N. benthamiana transcriptome using online databases (https://www.ncbi.nlm.nih.gov/).
3.2. HIGS-Assisted Screening for the Candidate Genes
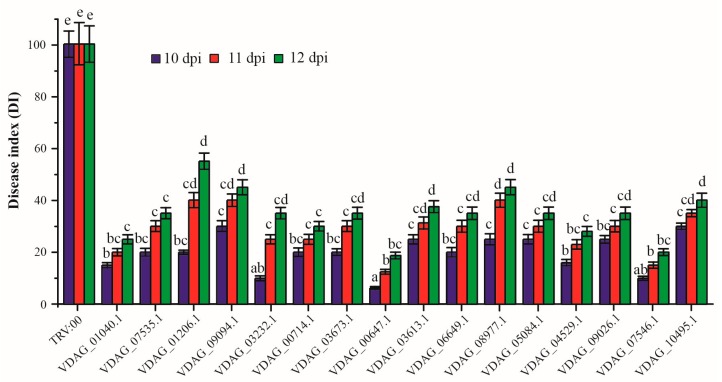
Ten days after infiltration, the seedlings were dipped into 106 spores/mL of resuspended Vd conidia, and the DI was calculated from severity scores at 10–12 dpi (Table S1). Plants that were injected with the TRV:00 (control group) showed typical wilting, stunting, chlorosis, and necrosis, and the DI reached 100 at 10 dpi. The plantlets in the RNAi groups, however, displayed varying levels of increased resistance to Vd compared with the control group.
The DI for the different RNAi groups gradually increased over time from 10 dpi. According to the Chinese national criteria for evaluating tolerance to Vw in cotton (GB/T 22101.5-2009), a plant variety is tolerant to Vw when DI <35. Within the RNAi groups, the DI for infected plants infiltrated with any of the 16 RNAi constructs used in the HIGS was obviously reduced (Table S6 and Figure 1). These 16 candidates are involved in energy metabolism, material transportation, protein modification, glucose metabolism, cell proliferation, DNA replication, and resistance. These results suggested that HIGS of Vd genes could be deployed to identify candidate virulence factors.
Figure 1.
Disease index over time for N. benthamiana seedlings infiltrated with 16 Vd strains that had been silenced for a candidate virulence-related gene.
3.3. VdAK May Be Responsible for Adaptation to Abiotic Stress
Based on the phenotypes and DI above, seedlings infiltrated with TRV:VdAK (VDAG_01040.1) displayed slight wilt. AK is a regulator of the metabolic pool of ATP, ADP, and AMP [46]. ADP and AMP are not only the source of energy [47], but also crucial factors in signal pathways for growth, development, and stress responses [48]. To ascertain the functions of VdAK, we generated disruption mutants (ΔVdAK) via homologous recombination, a GFP strain (ΔVdAK-GFP), and a complementation strain (ΔVdAK-C) via random insertion (Figure S2A–C). The transformants were selected after three generations and confirmed by RT-PCR (Figure S2D).
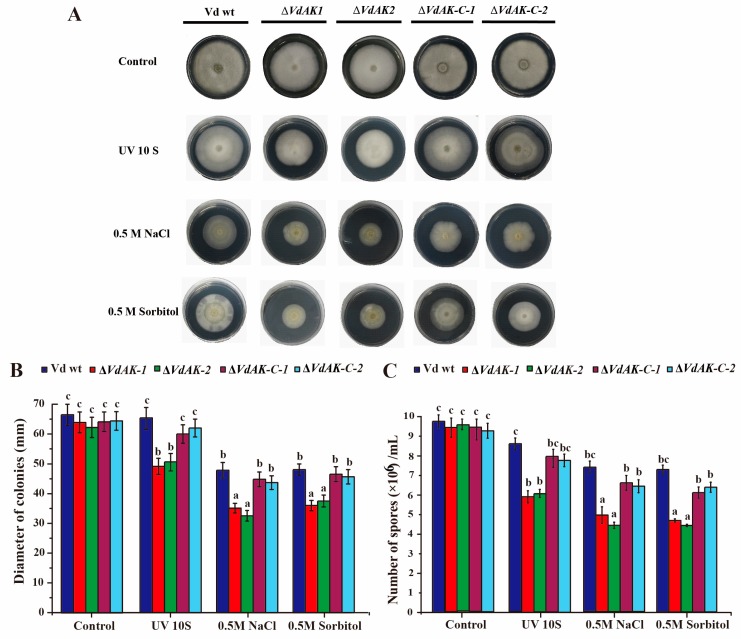
Subsequently, we investigated the putative roles of VdAK in adaptive responses of the fungus to various stresses. Strain ΔVdAK displayed no distinct defect in development or spore production, but growth and sporulation were significantly diminished by 0.5 M NaCl, 0.5 M sorbitol, and the 10 s UV treatment (Figure 2). Especially on media with NaCl and sorbitol, ΔVdAK colonies had statistically delayed hyphal growth and produced fewer spores than the wild-type Vd and ΔVdAK-C strains, which did not differ from each other. These results suggested VdAK has a positive role in fungal response to abiotic stress.
Figure 2.
The deletion of VdAK compromises mycelial growth and spore production during abiotic stress. A 10 µL drop of 106 spores/mL was placed in the center of Czapek Dox agar in a petri dish. Colony morphology (A), colony diameter (B), and spore production (C) of disruption (ΔVdAK) and complementation (ΔVdAK-C) mutants and wild-type Vd strain exposed to three abiotic stresses were analyzed after 4 weeks. Means (±SE) from three independent experiments were analyzed for significant differences among treatments using Duncan’s test (p <0.05). Different letters above bars within a treatment indicate significant differences among the strains.
3.4. VdAK Is Required for Full Vd Virulence
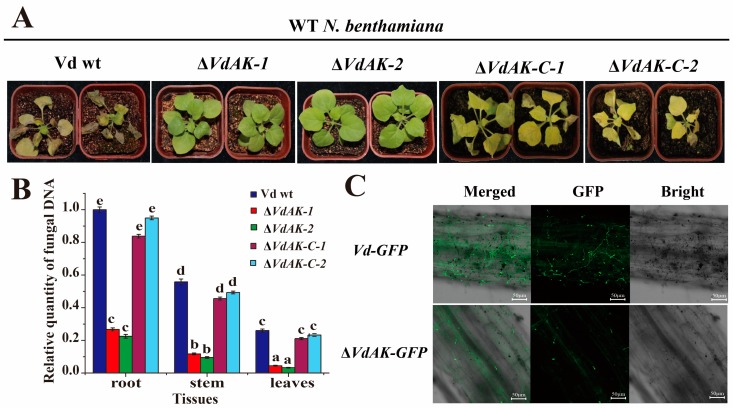
We further investigated the virulence of ΔVdAK, ΔVdAK-C, and wild-type Vd strains by inoculating 6-week-old seedlings of N. benthamiana with respective conidia. At 12 dpi, wilting symptoms and leaf necrosis were not visible on plants inoculated with ΔVdAK mutants, but symptoms were visible on plants inoculated with wild-type Vd (Figure 3A). The biomass of strain ΔVdAK was much lower in roots, stems, and leaves relative to that of the wild-type Vd (Figure 3B). Complementation strain ΔVdAK-C did not differ in virulence or biomass from the wild-type Vd (Figure 3A,B). Furthermore, hyphae of both Vd-GFP and ΔVdAK-GFP had colonized the roots by 5 dpi. Notably, spore germination was lower, and hyphae less abundant for the strain ΔVdAK-GFP than for Vd-GFP on the root surface (Figure 3C). In A. thaliana, strain ΔVdAK was less virulent and produced less biomass than did the wild-type and complementary strains, whereas the virulence of ΔVdAK-C and the wild-type Vd were equivalent (Figure S3A–C). Thus, these results confirmed that VdAK is indispensable for the full virulence of Vd.
Figure 3.
Virulence analysis of disruption (ΔVdAK), complementation (ΔVdAK-C), and wild-type Vd strains in N. benthamiana. (A) Symptoms on seedlings 12 days after roots were dipped in 106 spores/mL of ΔVdAK, ΔVdAK-C, or wild-type Vd (Vd wt). (B) Relative amounts of fungal DNA as determined by RT-qPCR. Means (± SE) from three independent experiments were analyzed for significant differences among treatments using Duncan’s test (p <0.05), as indicated by different letters. (C) Micrographs of fluorescing hyphae in N. benthamiana root tips at 5 dpi with Vd-GFP or ΔVdAK-GFP.
3.5. dsVdAK-Overexpressing Transgenic Plants Had Significant Resistance against Vd
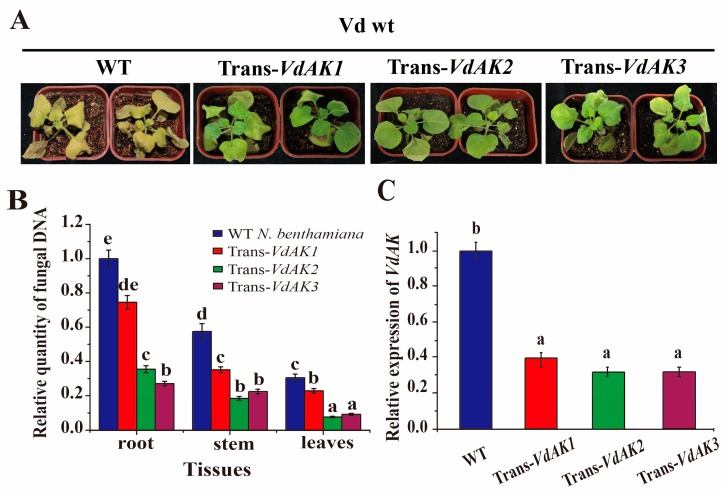
To further validate whether VdAK is associated with disease development, we generated stable transgenic lines of N. benthamiana and A. thaliana by inserting an RNAi construct with a 536 bp fragment of VdAK and the constitutive cauliflower mosaic viral 35S promoter. When 6-week-old T2 plants of three independent transgenic lines of N. benthamiana were tested for Vd resistance using the wild-type Vd, wilt symptoms were visibly milder at 12 dpi than on the control lines (Figure 4A). RT-qPCR analysis showed that the accumulation of fungal DNA was significantly suppressed in roots, stems, and leaves of the transgenic lines, relative to the wild-type (Figure 4B). The transcript level of VdAK in the transgenic lines was significantly reduced up to 4-fold compared with the wild-type (Figure 4C). In A. thaliana, the reduction of VdAK expression in transgenic plants resulted in increased Vd resistance and a reduction of fungal biomass (Figure S4A–C). Thus, the results demonstrated that the knockdown of VdAK expression compromised the virulence of Vd.
Figure 4.
Expression of dsRNA of VdAK improved Vd resistance in N. benthamiana compared to wild-type plants (WT). WT and trans-VdAK plants were inoculated with wild-type Vd (Vd wt). (A) Symptoms on transgenic and wild-type plants at 12 days after roots were dipped in 106 spores/mL of the respective strains. (B) Relative amounts of fungal DNA as determined by RT-qPCR. (C) Transcript levels of VdAK in stems of transgenic and WT plants. Means (± SE) from three independent experiments that differed significantly among treatments in Duncan’s test (p <0.05) are indicated by different letters.
3.6. VdAK Is Associated with Energy Metabolism
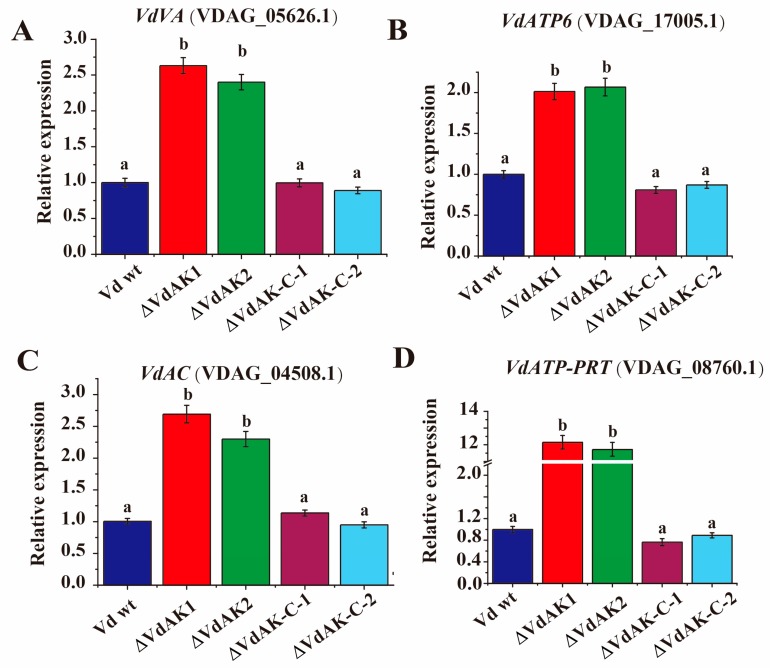
Among various proteins involved in energy metabolism, adenylate kinases have vital roles in maintaining an ADP/ATP balance [49,50]. In addition, the vacuolar ATPase (VA) can enhance vacuolar H+ pumping activity and Na+ compartmentalization capacity [51]. ATP6 is one of the three main subunits in membrane-localized ATP synthase F0, which is highly similar, functionally and mechanistically, to V-ATPase [52,53]. Furthermore, adenylate cyclase (AC), catalyzing the conversion of ATP to 3′,5′-cyclic AMP (cAMP), has key regulatory roles in signaling pathways [54]. ATP-phosphoribosyltransferase (ATP-PRT), the rate-limiting enzyme of the histidine pathway, is completely reversible depending on the ATP concentration [55,56]. To ascertain whether energy metabolism in Vd is affected with the knockout of VdAK, we quantified transcripts of the marker genes VdVA, VdATP6, VdAC and VdATP-PRT using RT-qPCR and harvested mycelium of strains ΔVdAK, ΔVdAK-C, and wild-type Vd, which had been cultured in CM broth for 7 days (Figure 5A–D). Transcript levels for VdVA, VdATP6, and VdAC were significantly elevated up to 2-fold, and VdATP-PRT transcripts increased significantly (>10-fold) in ΔVdAK when compared to levels in the ΔVdAK-C and wild-type Vd. Thus, manipulating the VdAK transcripts resulted in transcriptional reprogramming of these genes associated with energy metabolism.
Figure 5.
Relative expression of fungal genes associated with energy metabolism in ΔVdAK, ΔVdAK-C, and wild-type Vd (Vd wt) strains. Strains were cultured in complete medium (CM) broth for 7 days. Transcript levels of VdVA (A), VdATP6 (B), VdAC (C), and VdATP-PRT (D). Mean (± SE) levels from three independent experiments that differed significantly in Duncan’s test (p <0.05) are indicated by different letters.
3.7. A siRNA Targeting VdAK May Contribute to Cross-Kingdom Gene Silencing and Virulence Inhibition
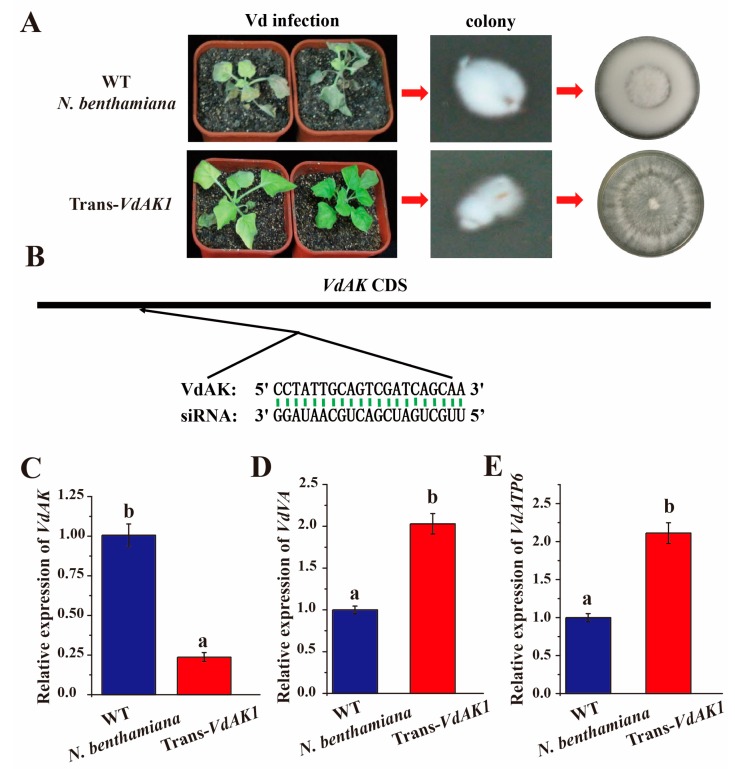
To further corroborate the reduction in wilt, the HIGS experiment was repeated in VdAK1-transgenic lines (Figure 6). At 12 dpi, the transgenic seedlings appeared to have significant resistance compared with WT N. benthamiana seedlings. Colonies recovered from transgenic seedling stems also differed distinctly from those from the WT seedlings (Figure 6A). To investigate whether siRNAs were generated in transgenic seedlings and entered Vd, the isolated fungi were cultured in CM broth, and the total RNA of the mycelium was subsequently extracted and sequenced (Figure 6B). Among small RNAs, one was found to target VdAK specifically (Figure 6B; Table S5). To validate HIGS, we further examined the VdAK transcripts in Vd using RT-qPCR. Transcript levels for VdAK were downregulated about 4-fold by HIGS (Figure 6C). Moreover, the expression of two marker genes, determined using RT-qPCR, was upregulated after VdAK silencing (Figure 6D,E). These results support the view that exogenous siRNAs can enter pathogenic fungi through plants and silence the target gene to improve host resistance.
Figure 6.
Host-induced gene silencing of VdAK enhanced resistance against V. dahliae (Vd). (A) Disease symptoms on N. benthamiana at 12 dpi and growth phenotype on PDA of Vd isolated from infected seedlings. (B) Specific siRNA targeted the VdAK position. The siRNA sequence aligned with VdAK at the predicted binding site. Relative expression levels of VdAK (C), VdVA (D), and VdATP6 (E) in RT-qPCR analysis of the recovered mycelium. Mean (± SE) from three independent experiments that differed significantly in Duncan’s test (p <0.05) are indicated by different letters.
4. Discussion
In this study, we first developed a HIGS system for large-scale screening of pathogenicity/virulence factors, and found 16 potential candidates. Subsequently, we confirmed that the VdAK plays a vital role in fungal metabolism, conidiation, and pathogenicity and could be a valuable gene to transform plants for increased resistance to Vd. Indeed, the RNAi-VdAK transgenic plants exhibited significant resistance to Vd. Notably, siRNA that targeted VdAK was identified in transgenic plants. It may prove to be useful for cross-kingdom gene silencing.
In eukaryotic organisms, RNA interference (RNAi) is a highly conserved mechanism, and a valid tool to knock down gene expression [57,58,59]. Based on this mechanism, the TRV-mediated gene silencing has been used to downregulate endogenous genes in plants [36,38,60]. Recently, HIGS has also proved to be a promising strategy for silencing a foreign gene in a host to investigate numerous plant–pathogen systems [61,62], including Vd in various hosts [33,34,63]. In the present study, positive plants at 10 days after infiltration showed distinct photobleaching, suggesting the massive presence of dsRNA and its active role in gene silencing [64,65,66]. In our study, seedlings that were inoculated with Vd developed varying degrees of wilting from 10 to 12 dpi. The whole process took about 25 days. Hence, HIGS can be utilized as an efficient platform for genome-wide high-throughput RNAi screening to identify pathogenic genes. We also screened for Vd candidate pathogenicity/virulence factor genes using publicly available genomic resources and the TRV system. Within a comparatively short time, we identified 16 candidate genes, that when silenced, resulted in significant resistance to Vd.
We used a GO analysis for our preliminary selection of candidate genes of Vd involved in highly conserved biological function. A similar approach has also been used for Blumeria graminis in which targeted silencing of important genes such as heat shock protein 70, 40S ribosomal protein, and ADP/ATP carrier protein led to reduced sporulation [26]. We focused on an in-depth analysis of one candidate gene, VdAK, that encodes a putative adenylate kinase, whose reduced expression led to an obvious decrease in the virulence of Vd. Adenylate kinase is a critical phosphotransferase that catalyzes the interconversion of ADP and ATP [67,68,69]. Transphosphorylation of adenine nucleotides regulates the cellular concentrations of ADP, ATP, and AMP, which directly affects the adenylate distribution during glucose and nucleic acid metabolism [70,71]. This gene is indispensable for controlling the balance between ATP and ADP in cells [67,72] and thus is important for adaptation to diverse stresses [73], as shown by our analysis of ΔVdAK strains exposed to various stresses. Under stress, strain ΔVdAK grew significantly less and produced fewer spores compared to the wild-type. This decrease was rescued by complementation with a functional VdAK gene. Using the VdAK disruption and VdAK complementation mutant strains, we found that disruption of VdAK increased fungal sensitivity to different stresses. Thus, VdAK apparently contributes to improved responses to diverse stresses.
Transcripts of VdAC, VdATP6, VdAC, and VdATP-PRT were upregulated in strain ΔVdAK, presumably triggered by a high ratio of ADP/AMP [74]. The upregulation of the vacuolar ATPase (VA) gene is associated with improved salt tolerance in halotolerant peppermint Keyuan-1 [51]. The upregulation of VdVA, VdATP6, and VdAC might also be related to ATP synthesis and the signal network [75,76,77]. When VdATP-PRT expression is strongly enhanced, glycometabolism may be stimulated and thus provide cellular energy [78,79,80]. Taken together, the accumulative evidence suggests that VdAK is a positive virulence regulator that is linked to energy metabolism. Moreover, the ability to germinate and colonize the root surface of N. benthamiana and A. thaliana may also be impaired in ΔVdAK, leading to less colonization in the plant vessels. Our results clearly indicate that the VdAK gene positively regulates virulence of Vd. Given the roles of AK in energy metabolism, when VdAK is suppressed, ADP/ATP turnover might also be disturbed, reducing fungal growth, development, and virulence. The compromised virulence and stress tolerance of the ΔVdAK disruption mutant support this hypothesis.
As a group of small, noncoding RNAs, siRNAs regulate post-transcriptional gene expression and participate in diverse biological processes, including resistance against stress [81,82,83,84]. In screening and study of a class of miRNAs related to Phytophthora sojae resistance in three soybean cultivars [85], the expression of miR393 in soybean significantly increases in response to P. sojae infection [86]. After Vd infection, cotton and A. thaliana increase the production of miRNA166 and miRNA159 to target Vd genes and result in improved resistance [87]. Five miRNAs in a highly resistant strain of Vitis davidii, revealed by microRNA sequencing, were specifically expressed and used to investigate further the potential inhibition of grape white rot disease caused by Coniella diplodiwlla [88]. In our study, a specific siRNA against VdAK was detected in Vd isolated from transgenic plants, which provides further insights into the action between siRNA and fungi in planta. VdAK was silenced due to the presence of siRNA, which results in the increased expression of VdVA and VdATP6 involved in energy metabolism. These results are consistent with the expression of the marker genes in ΔVdAK strains. The reduced wilt symptoms are likely caused by the disruption of energy metabolism of Vd and subsequent growth in transgenic seedlings. These results illustrate that the VdAK gene has potential as another target for a HIGS strategy to control Vd.
5. Conclusions
In summary, we confirmed that the HIGS system is very efficient for screening candidate pathogenic factors in Vd. As a positive regulator needed for full virulence, VdAK holds promise as a target to enhance the resistance of transgenic plants harboring dsVdAK against Vd.
Acknowledgments
The authors thank Yule Liu (Tsinghua University) for providing the HIGS plasmids, Xiaofeng Dai (Institute of Food Science and Technology, CAAS) for providing the Vd-GFP strain, and Guiliang Jian (Institute of Crop Protection, CAAS) for providing Vd strain V991.
Supplementary Materials
The supplementary materials are available online at https://www.mdpi.com/2218-273X/10/1/127/s1.
Author Contributions
H.C., G.W., and H.G. conceived and designed the experiments. X.S., G.L., X.L., and L.R. performed experiments and analyzed data. X.S., W.L., and G.S. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by a grant from the National Transgenic Major Project of China (2019ZX08010-004), the National Natural Science Foundation of China (31701861 and 31772244), the special fund for agro-scientific research in the public interest (201503109), the Agricultural Science and Technology Innovation Program of CAAS, the Fundamental Research Funds for Central Non-profit Scientific Institution (Y2017JC57), and the Project of the Innovation Team Building in Key Areas of Xinjiang Production and Construction Corps (XPCC) (2019CB008).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Fradin E.F., Thomma B.P. Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Mol. Plant Pathol. 2006;7:71–86. doi: 10.1111/j.1364-3703.2006.00323.x. [DOI] [PubMed] [Google Scholar]
- 2.Bhat R.G., Subbarao K.V. Host range specificity in Verticillium dahliae. Phytopathology. 1999;89:1218–1525. doi: 10.1094/PHYTO.1999.89.12.1218. [DOI] [PubMed] [Google Scholar]
- 3.Barbara D.J., Clewes E. Plant pathogenic Verticillium species: How many of them are there? Mol. Plant Pathol. 2003;4:297–305. doi: 10.1046/j.1364-3703.2003.00172.x. [DOI] [PubMed] [Google Scholar]
- 4.Klosterman S.J., Atallah Z.K., Vallad G.E., Subbarao K.V. Diversity, pathogenicity, and management of Verticillium species. Annu. Rev. Phytopathol. 2009;47:39–62. doi: 10.1146/annurev-phyto-080508-081748. [DOI] [PubMed] [Google Scholar]
- 5.Li C.H., Feng Z.L., Li Z.F., Zhang Z.G., Shi Y.Q., Zhao L.H., Zhu H.Q. In vitro sensitivity of Verticillium dahliae Kleb strains against some effective fungicides. China Cotton. 2015;42:16–18. [Google Scholar]
- 6.Klosterman S.J., Subbarao K.V., Kang S., Veronese P., Gold S.E., Thomma B.P., Chen Z., Henrissat B., Lee Y.H., Park J., et al. Comparative genomics yields insights into niche adaptation of plant vascular wilt pathogens. PLoS Pathog. 2011;7:e1002137. doi: 10.1371/journal.ppat.1002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang X., Ben S., Sun Y., Fan X., Tian C., Wang Y. Genome-wide identification, phylogeny and expression profile of vesicle fusion components in Verticillium dahliae. PLoS ONE. 2013;8:e68681. doi: 10.1371/journal.pone.0068681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duressa D., Anchieta A., Chen D., Klimes A., Garcia-Pedrajas M.D., Dobinson K.F., Klosterman S.J. RNA-seq analyses of gene expression in the microsclerotia of Verticillium dahliae. BMC Genomics. 2013;14:498. doi: 10.1186/1471-2164-14-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hadwiger L.A., Druffel K., Humann J.L., Schroeder B.K. Nuclease released by Verticillium dahliae is a signal for non-host resistance. Plant Sci. 2013;201:98–107. doi: 10.1016/j.plantsci.2012.11.011. [DOI] [PubMed] [Google Scholar]
- 10.Tran V.T., Braus-Stromeyer S.A., Kusch H., Reusche M., Kaever A., Kuhn A., Valerius O., Landesfeind M., Asshauer K., Tech M., et al. Verticillium transcription activator of adhesion Vta2 suppresses microsclerotia formation and is required for systemic infection of plant roots. New Phytol. 2014;202:565–581. doi: 10.1111/nph.12671. [DOI] [PubMed] [Google Scholar]
- 11.Tzima A.K., Paplomatas E.J., Rauyaree P., Ospina-Giraldo M.D., Kang S. VdSNF1, the sucrose nonfermenting protein kinase gene of Verticillium dahliae, is required for virulence and expression of genes involved in cell-wall degradation. Mol. Plant Microbe Interact. 2011;24:129–142. doi: 10.1094/MPMI-09-09-0217. [DOI] [PubMed] [Google Scholar]
- 12.Tzima A.K., Paplomatas E.J., Tsitsigiannis D.I., Kang S. The G protein beta subunit controls virulence and multiple growth- and development-related traits in Verticillium dahliae. Fungal Genet. Biol. 2012;49:271–283. doi: 10.1016/j.fgb.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 13.Eboigbe L., Tzima A.K., Paplomatas E.J., Typas M.A. The role of the β-1, 6-endoglucanase gene vegB in physiology and virulence of Verticillium dahliae. Phytopathol. Mediterr. 2014;53:94–107. [Google Scholar]
- 14.Tian L., Xu J., Zhou L., Guo W. VdMsb regulates virulence and microsclerotia production in the fungal plant pathogen Verticillium dahliae. Gene. 2014;550:238–244. doi: 10.1016/j.gene.2014.08.035. [DOI] [PubMed] [Google Scholar]
- 15.Xiong D., Wang Y., Tian L., Tian C. MADS-Box transcription factor VdMcm1 regulates conidiation, microsclerotia formation, pathogenicity, and secondary metabolism of Verticillium dahliae. Front. Microbiol. 2016;7:1192–1206. doi: 10.3389/fmicb.2016.01192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng Z., Tian J., Han L., Geng Y., Sun J., Kong Z. The Myosin5-mediated actomyosin motility system is required for Verticillium pathogenesis of cotton. Environ. Microbiol. 2018;20:1607–1621. doi: 10.1111/1462-2920.14101. [DOI] [PubMed] [Google Scholar]
- 17.Bu B., Qiu D., Zeng H., Guo L., Yuan J., Yang X. A fungal protein elicitor PevD1 induces Verticillium wilt resistance in cotton. Plant Cell Rep. 2014;33:461–470. doi: 10.1007/s00299-013-1546-7. [DOI] [PubMed] [Google Scholar]
- 18.Xie C., Li Q., Yang X. Characterization of VdASP F2 secretory factor from Verticillium dahliae by a fast and easy gene knockout system. Mol. Plant Microbe Interact. 2017;30:444–454. doi: 10.1094/MPMI-01-17-0007-R. [DOI] [PubMed] [Google Scholar]
- 19.Bui T.T., Harting R., Braus-Stromeyer S.A., Tran V.T., Leonard M., Hofer A., Abelmann A., Bakti F., Valerius O., Schluter R., et al. Verticillium dahliae transcription factors Som1 and Vta3 control microsclerotia formation and sequential steps of plant root penetration and colonisation to induce disease. New Phytol. 2019;221:2138–2159. doi: 10.1111/nph.15514. [DOI] [PubMed] [Google Scholar]
- 20.Zhang W.Q., Gui Y.J., Short D.P.G., Li T.G., Zhang D.D., Zhou L., Liu C., Bao Y.M., Subbarao K.V., Chen J.Y., et al. Verticillium dahliae transcription factor VdFTF1 regulates the expression of multiple secreted virulence factors and is required for full virulence in cotton. Mol. Plant Pathol. 2018;19:841–857. doi: 10.1111/mpp.12569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tinoco M.L., Dias B.B., Dall’Astta R.C., Pamphile J.A., Aragao F.J. In vivo trans-specific gene silencing in fungal cells by in planta expression of a double-stranded RNA. BMC Biol. 2010;8:27–37. doi: 10.1186/1741-7007-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baulcombe D.C. VIGS, HIGS and FIGS: Small RNA silencing in the interactions of viruses or filamentous organisms with their plant hosts. Curr. Opin. Plant Biol. 2015;26:141–146. doi: 10.1016/j.pbi.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 23.Amberkar S., Kiani N.A., Bartenschlager R., Alvisi G., Kaderali L. High-throughput RNA interference screens integrative analysis: Towards a comprehensive understanding of the virus-host interplay. World J. Virol. 2013;2:18–31. doi: 10.5501/wjv.v2.i2.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hua C., Zhao J.H., Guo H.S. Trans-kingdom RNA silencing in plant-fungal pathogen interactions. Mol. Plant. 2018;11:235–244. doi: 10.1016/j.molp.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 25.Nunes C.C., Dean R.A. Host-induced gene silencing: A tool for understanding fungal host interaction and for developing novel disease control strategies. Mol. Plant Pathol. 2012;13:519–529. doi: 10.1111/j.1364-3703.2011.00766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nowara D., Gay A., Lacomme C., Shaw J., Ridout C., Douchkov D., Hensel G., Kumlehn J., Schweizer P. HIGS: Host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell. 2010;22:3130–3141. doi: 10.1105/tpc.110.077040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Panwar V., McCallum B., Bakkeren G. Host-induced gene silencing of wheat leaf rust fungus Puccinia triticina pathogenicity genes mediated by the Barley stripe mosaic virus. Plant Mol. Biol. 2013;81:595–608. doi: 10.1007/s11103-013-0022-7. [DOI] [PubMed] [Google Scholar]
- 28.Mumbanza F.M., Kiggundu A., Tusiime G., Tushemereirwe W.K., Niblett C., Bailey A. In vitro antifungal activity of synthetic dsRNA molecules against two pathogens of banana, Fusarium oxysporum f. sp. cubense and Mycosphaerella fijiensis. Pest Manage. Sci. 2013;69:1155–1162. doi: 10.1002/ps.3480. [DOI] [PubMed] [Google Scholar]
- 29.Ghag S.B., Shekhawat U.K., Ganapathi T.R. Host-induced post-transcriptional hairpin RNA-mediated gene silencing of vital fungal genes confers efficient resistance against Fusarium wilt in banana. Plant Biotechnol. J. 2014;12:541–553. doi: 10.1111/pbi.12158. [DOI] [PubMed] [Google Scholar]
- 30.Chen W., Kastner C., Nowara D., Oliveira-Garcia E., Rutten T., Zhao Y., Deising H.B., Kumlehn J., Schweizer P. Host-induced silencing of Fusarium culmorum genes protects wheat from infection. J. Exp. Bot. 2016;67:4979–4991. doi: 10.1093/jxb/erw263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu J., Wang X., Li Y., Zeng J., Wang G., Deng C., Guo W. Host-induced gene silencing of a regulator of G protein signalling gene (VdRGS1) confers resistance to Verticillium wilt in cotton. Plant Biotechnol. J. 2018;16:1629–1643. doi: 10.1111/pbi.12900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song Y., Thomma B. Host-induced gene silencing compromises Verticillium wilt in tomato and Arabidopsis. Mol. Plant Pathol. 2018;19:77–89. doi: 10.1111/mpp.12500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su X., Rehman L., Guo H., Li X., Cheng H. The oligosaccharyl transferase subunit STT3 mediates fungal development and is required for virulence in Verticillium dahliae. Curr. Genet. 2018;64:235–246. doi: 10.1007/s00294-017-0729-0. [DOI] [PubMed] [Google Scholar]
- 34.Su X., Rehman L., Guo H., Li X., Zhang R., Cheng H. AAC as a potential target gene to control Verticillium dahliae. Genes. 2017;8:25–41. doi: 10.3390/genes8010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Z., Song Y., Liu C.M., Thomma B.P. Mutational analysis of the Ve1 immune receptor that mediates Verticillium resistance in tomato. PLoS ONE. 2014;9:e99511. doi: 10.1371/journal.pone.0099511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y., Schiff M., Dinesh-Kumar S.P. Virus-induced gene silencing in tomato. Plant J. 2002;31:777–786. doi: 10.1046/j.1365-313X.2002.01394.x. [DOI] [PubMed] [Google Scholar]
- 37.Hayward A., Padmanabhan M., Dinesh-Kumar S.P. Virus-induced gene silencing in Nicotiana benthamiana and other plant species. Methods Mol. Biol. 2011;678:55–63. doi: 10.1007/978-1-60761-682-5_5. [DOI] [PubMed] [Google Scholar]
- 38.Zhou Y., Sun L., Wassan G.M., He X., Shaban M., Zhang L., Zhu L., Zhang X. GbSOBIR1 confers Verticillium wilt resistance by phosphorylating the transcriptional factor GbbHLH171 in Gossypium barbadense. Plant Biotechnol. J. 2019;17:152–163. doi: 10.1111/pbi.12954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clough S.J., Bent A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 40.Mao Y., Cai W., Wang J., Hong G., Tao X., Wang L., Huang Y., Chen X. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat. Biotechnol. 2007;25:1307–1313. doi: 10.1038/nbt1352. [DOI] [PubMed] [Google Scholar]
- 41.Wang H.M., Lin Z.X., Zhang X.L., Chen W., Guo X.P., Nie Y.C., Li Y.H. Mapping and quantitative trait loci analysis of Verticillium wilt resistance genes in cotton. J. Integr. Plant Biol. 2008;50:174–182. doi: 10.1111/j.1744-7909.2007.00612.x. [DOI] [PubMed] [Google Scholar]
- 42.Rehman L., Su X., Guo H., Qi X., Cheng H. Protoplast transformation as a potential platform for exploring gene function in Verticillium dahliae. BMC Biotechnol. 2016;16:57–65. doi: 10.1186/s12896-016-0287-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoppenau C.E., Tran V.-T., Kusch H., Aßhauer K.P., Landesfeind M., Meinicke P., Popova B., Braus-Stromeyer S.A., Braus G.H. Verticillium dahliae VdTHI4, involved in thiazole biosynthesis, stress response and DNA repair functions, is required for vascular disease induction in tomato. Environ. Exp. Bot. 2014;108:14–22. doi: 10.1016/j.envexpbot.2013.12.015. [DOI] [Google Scholar]
- 44.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 45.Zhao Y., Su X., Cheng H. Verification of Verticillium dahliae pathogenicity of glycometabolism related genes by using host-induced gene silencing method. Sci. Gric. Sin. 2015;48:1321–1329. [Google Scholar]
- 46.Noma T. Dynamics of nucleotide metabolism as a supporter of life phenomena. J. Investig. Med. 2005;52:127–136. doi: 10.2152/jmi.52.127. [DOI] [PubMed] [Google Scholar]
- 47.Schulz G. Structural and Functional Relationships in the Adenylate Kinase Family. Cold Spring Harb. Symp. Quant. Boil. 1987;52:429–439. doi: 10.1101/SQB.1987.052.01.050. [DOI] [PubMed] [Google Scholar]
- 48.Hardie D.G., Carling D. The AMP-activated protein kinase fuel gauge of the mammalian cell? Eur. J. Biochem. 1997;246:259–273. doi: 10.1111/j.1432-1033.1997.00259.x. [DOI] [PubMed] [Google Scholar]
- 49.Miura K., Inouye S., Sakai K., Takaoka H., Kishi F., Tabuchi M., Tanaka T., Matsumoto H., Shirai M., Nakazawa T., et al. Cloning and characterization of adenylate kinase from Chlamydia pneumoniae. J. Biol. Chem. 2001;276:13490–13498. doi: 10.1074/jbc.M009461200. [DOI] [PubMed] [Google Scholar]
- 50.Claypool S.M., Oktay Y., Boontheung P., Loo J.A., Koehler C.M. Cardiolipin defines the interactome of the major ADP/ATP carrier protein of the mitochondrial inner membrane. J. Cell Biol. 2008;182:937–950. doi: 10.1083/jcb.200801152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Z., Zhen Z., Guo K., Harvey P., Li J., Yang H. MAPK-mediated enhanced expression of vacuolar H-ATPase confers the improved adaption to NaCl stress in a halotolerate peppermint (Mentha piperita L.) Protoplasma. 2015;253:553–569. doi: 10.1007/s00709-015-0834-1. [DOI] [PubMed] [Google Scholar]
- 52.McCarty R.E. A plant biochemist’s view of H+-ATPases and ATP synthases. J. Exp. Biol. 1992;172:431–441. doi: 10.1242/jeb.172.1.431. [DOI] [PubMed] [Google Scholar]
- 53.Kim M.S., Jang J., Ab Rahman N.B., Pethe K., Berry E.A., Huang L.S. Isolation and characterization of a hybrid respiratory supercomplex consisting of Mycobacterium tuberculosis cytochrome bcc and Mycobacterium smegmatis cytochrome aa3. J. Biol. Chem. 2015;290:14350–14360. doi: 10.1074/jbc.M114.624312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Salomon Y., Londos C., Rodbell M. A highly sensitive adenylate cyclase assay. Anal. Biochem. 1974;58:541–548. doi: 10.1016/0003-2697(74)90222-X. [DOI] [PubMed] [Google Scholar]
- 55.Alifano P., Fani R. Histidine biosynthetic pathway and genes: Structure, regulation, and evolution. Microbiol. Rev. 1996;60:44–69. doi: 10.1128/MMBR.60.1.44-69.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blasi F., Aloj S.M., Goldberger R.F. Effect of histidine on the enzyme which catalyzes the first step of histidine biosynthesis in Salmonella typhimurium. Biochemistry. 1971;10:1409–1417. doi: 10.1021/bi00784a021. [DOI] [PubMed] [Google Scholar]
- 57.Clemens J.C., Worby C.A., Simonson-Leff N., Muda M., Maehama T., Hemmings B.A., Dixon J.E. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Natl. Acad. Sci. USA. 2000;97:6499–6503. doi: 10.1073/pnas.110149597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baulcombe D. RNA silencing. Trends Biochem. Sci. 2005;30:290–293. doi: 10.1016/j.tibs.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 59.Duan C.G., Wang C.H., Guo H.S. Application of RNA silencing to plant disease resistance. Silence. 2012;3:5–12. doi: 10.1186/1758-907X-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deng-wei J., Liu Y., Ce S., Min C., Qing Y. Cloning and characterization of a Solanum torvum NPR1 gene involved in regulating plant resistance to Verticillium dahliae. Acta Physiol. Plant. 2014;36:2999–3011. doi: 10.1007/s11738-014-1671-0. [DOI] [Google Scholar]
- 61.Helber N., Wippel K., Sauer N., Schaarschmidt S., Hause B., Requena N. A versatile monosaccharide transporter that operates in the arbuscular mycorrhizal fungus Glomus sp is crucial for the symbiotic relationship with plants. Plant Cell. 2011;23:3812–3823. doi: 10.1105/tpc.111.089813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jahan S.N., Asman A.K., Corcoran P., Fogelqvist J., Vetukuri R.R., Dixelius C. Plant-mediated gene silencing restricts growth of the potato late blight pathogen Phytophthora infestans. J. Exp. Bot. 2015;66:2785–2794. doi: 10.1093/jxb/erv094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang T., Jin Y., Zhao J.H., Gao F., Zhou B.J., Fang Y.Y., Guo H.S. Host-induced gene silencing of target gene in fungal cells confers effective resistance to cotton wilt disease pathogen Verticillium dahliae. Mol. Plant. 2016;9:939–942. doi: 10.1016/j.molp.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 64.Pang J., Zhu Y., Li Q., Liu J., Tian Y., Liu Y., Wu J. Development of Agrobacterium-mediated virus-induced gene silencing and performance evaluation of four marker genes in Gossypium barbadense. PLoS One. 2013;8:e73211. doi: 10.1371/journal.pone.0073211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu H., Xu L., Yang P., Cao Y., Tang Y., He G., Yuan S., Ming J. Tobacco rattle virus-induced PHYTOENE DESATURASE (PDS) and Mg-chelatase H subunit (ChlH) gene silencing in Solanum pseudocapsicum L. PeerJ. 2018;6:e4424. doi: 10.7717/peerj.4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang J., Yu D., Zhang Y., Liu K., Xu K., Zhang F., Wang J., Tan G., Nie X., Ji Q., et al. Vacuum and co-cultivation Agroinfiltration of (germinated) seeds results in tobacco rattle virus (TRV) mediated whole-plant virus-induced gene silencing (VIGS) in wheat and maize. Front. Plant Sci. 2017;8:393–404. doi: 10.3389/fpls.2017.00393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Noda L. 8 Adenylate Kinase. The Enzymes. 1973;8:279–305. [Google Scholar]
- 68.Wang Y., Makowski L. Fine structure of conformational ensembles in adenylate kinase. Proteins. 2018;86:332–343. doi: 10.1002/prot.25443. [DOI] [PubMed] [Google Scholar]
- 69.Pradet A., Raymond P. Adenine nucleotide ratios and adenylate energy charge in energy metabolism. Ann. Rev. Plant Phys. 1983;34:199–224. doi: 10.1146/annurev.pp.34.060183.001215. [DOI] [Google Scholar]
- 70.Atkinson D.E. The energy charge of the adenylate pool as a regulatory parameter. Interaction with feedback modifiers. Biochemistry. 1968;7:4030–4034. doi: 10.1021/bi00851a033. [DOI] [PubMed] [Google Scholar]
- 71.Gauthier S., Coulpier F., Jourdren L., Merle M., Beck S., Konrad M., Daignan-Fornier B., Pinson B. Co-regulation of yeast purine and phosphate pathways in response to adenylic nucleotide variations. Mol. Microbiol. 2008;68:1583–1594. doi: 10.1111/j.1365-2958.2008.06261.x. [DOI] [PubMed] [Google Scholar]
- 72.Dzeja P., Terzic A. Adenylate kinase and AMP signaling networks: Metabolic monitoring, signal communication and body energy sensing. Int. J. Mol. Sci. 2009;10:1729–1772. doi: 10.3390/ijms10041729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dzeja P.P., Vitkevicius K.T., Redfield M.M., Burnett J.C., Terzic A. Adenylate kinase-catalyzed phosphotransfer in the myocardium: Increased contribution in heart failure. Circ. Res. 1999;84:1137–1143. doi: 10.1161/01.RES.84.10.1137. [DOI] [PubMed] [Google Scholar]
- 74.Bellinzoni M., Haouz A., Grana M., Munier-Lehmann H., Shepard W., Alzari P.M. The crystal structure of Mycobacterium tuberculosis adenylate kinase in complex with two molecules of ADP and Mg2+ supports an associative mechanism for phosphoryl transfer. Protein Sci. 2006;15:1489–1493. doi: 10.1110/ps.062163406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sun-Wada G.H., Wada Y. Role of vacuolar-type proton ATPase in signal transduction. Biochim. Biophys. Acta. 2015;1847:1166–1172. doi: 10.1016/j.bbabio.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 76.Manfredi G., Fu J., Ojaimi J., Sadlock J.E., Kwong J.Q., Guy J., Schon E.A. Rescue of a deficiency in ATP synthesis by transfer of MTATP6, a mitochondrial DNA-encoded gene, to the nucleus. Nat. Genet. 2002;30:394–399. doi: 10.1038/ng851. [DOI] [PubMed] [Google Scholar]
- 77.Bassler J., Schultz J.E., Lupas A.N. Adenylate cyclases: Receivers, transducers, and generators of signals. Cell. Signal. 2018;46:135–144. doi: 10.1016/j.cellsig.2018.03.002. [DOI] [PubMed] [Google Scholar]
- 78.Mittelstadt G., Jiao W., Livingstone E.K., Moggre G.J., Nazmi A.R., Parker E.J. A dimeric catalytic core relates the short and long forms of ATP-phosphoribosyltransferase. Biochem. J. 2018;475:247–260. doi: 10.1042/BCJ20170762. [DOI] [PubMed] [Google Scholar]
- 79.Livingstone E.K., Mittelstadt G., Given F.M., Parker E.J. Independent catalysis of the short form HisG from Lactococcus lactis. FEBS Lett. 2016;590:2603–2610. doi: 10.1002/1873-3468.12277. [DOI] [PubMed] [Google Scholar]
- 80.Champagne K.S., Piscitelli E., Francklyn C.S. Substrate recognition by the hetero-octameric ATP phosphoribosyltransferase from Lactococcus lactis. Biochemistry. 2006;45:14933–14943. doi: 10.1021/bi061802v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guarnieri D.J., DiLeone R.J. MicroRNAs: A new class of gene regulators. Ann. Med. 2008;40:197–208. doi: 10.1080/07853890701771823. [DOI] [PubMed] [Google Scholar]
- 82.Diederichs S., Haber D.A. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131:1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 83.Chen Y., Dong J., Bennetzen J.L., Zhong M., Yang J., Zhang J., Li S., Hao X., Zhang Z., Wang X. Integrating transcriptome and microRNA analysis identifies genes and microRNAs for AHO-induced systemic acquired resistance in N. tabacum. Sci. Rep. 2017;7:12504–12516. doi: 10.1038/s41598-017-12249-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cui L.G., Shan J.X., Shi M., Gao J.P., Lin H.X. The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J. 2015;80:1108–1117. doi: 10.1111/tpj.12712. [DOI] [PubMed] [Google Scholar]
- 85.Guo N., Ye W.W., Wu X.L., Shen D.Y., Wang Y.C., Xing H., Dou D.L. Microarray profiling reveals microRNAs involving soybean resistance to Phytophthora sojae. Genome. 2011;54:954–958. doi: 10.1139/g11-050. [DOI] [PubMed] [Google Scholar]
- 86.Wong J., Gao L., Yang Y., Zhai J., Arikit S., Yu Y., Duan S., Chan V., Xiong Q., Yan J., et al. Roles of small RNAs in soybean defense against Phytophthora sojae infection. Plant J. 2014;79:928–940. doi: 10.1111/tpj.12590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang T., Zhao Y.L., Zhao J.H., Wang S., Jin Y., Chen Z.Q., Fang Y.Y., Hua C.L., Ding S.W., Guo H.S. Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nat. Plants. 2016;2:16153–16158. doi: 10.1038/nplants.2016.153. [DOI] [PubMed] [Google Scholar]
- 88.Zhang Y., Fan X., Jiang J., Li M., Liu C. Analysis of the function of miRNA on the resisitance to white-rot disease in Vitis davidii based on microRNA sequencing. J. Fruit Sci. 2019;36:143–152. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.