Fig. 1. Biogenesis of circRNAs.
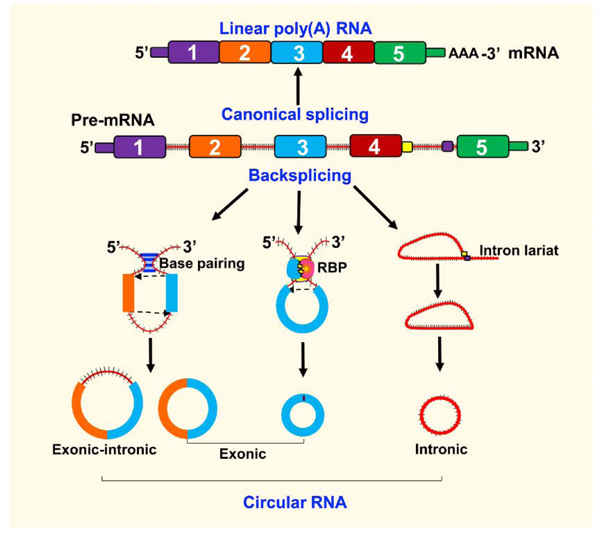
CircRNAs are formed from either exons, introns or both exon-intron by back-splicing events and spliceosomal machinery. On the contrary, canonical splicing forms a mature mRNA after removal of introns. Various processes that form circRNAs are conventional back-splicing driven, intron-pairing-driven circularization, and lariat-driven circularization. The downstream donor and upstream acceptor sites are brought into close proximity during circularization by exon-containing lariats, direct base pairing between cis-acting regulatory elements containing reverse complementary sequences (Alu repeats) and flanking introns, trans-acting factors, such as RNA-binding proteins (RBPs, QKI, MBL) and intron lariats that escape the usual intron debranching and degradation.
