Fig. 2. Methods to detect circRNAs.
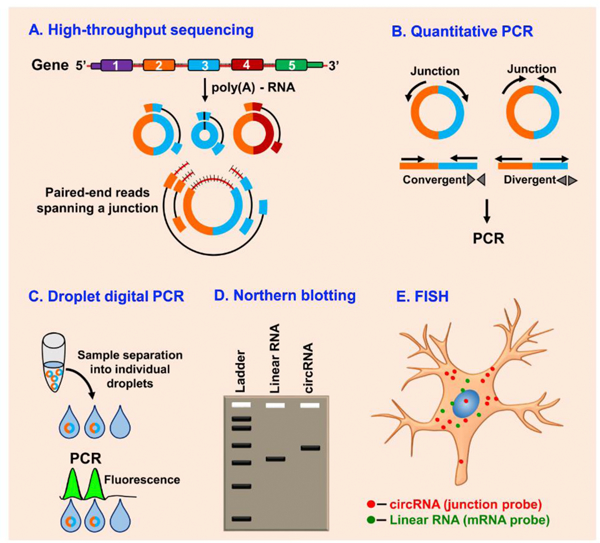
Coupled with the bioinformatics algorithms, deep sequencing and longer reads of RNAs digested with RNase R and poly(A) depleted RNAs increases the specificity of circRNAs detection (A). PCR analysis with convergent and divergent primers in RNase R-treated RNA preparations can be used to detect circRNAs (B). Droplet digital PCR quantifies absolute circRNA levels based on the single droplet molecules (C). Northern blotting can accurately detect and confirm an exact circRNA in a gel using probes that are designed to target both the circular and the linear transcript or only the circRNA with backsplice junction probe (D). Finally, RNA fluorescence in situ hybridization (RNA-FISH) coupled with high-resolution microscopy using probes flanking the junction sites can determine the distribution and abundance of circRNAs (E).
