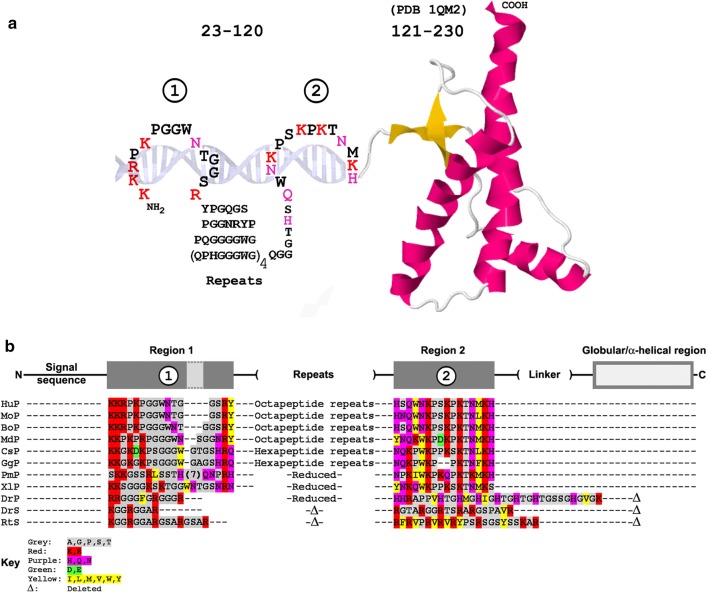
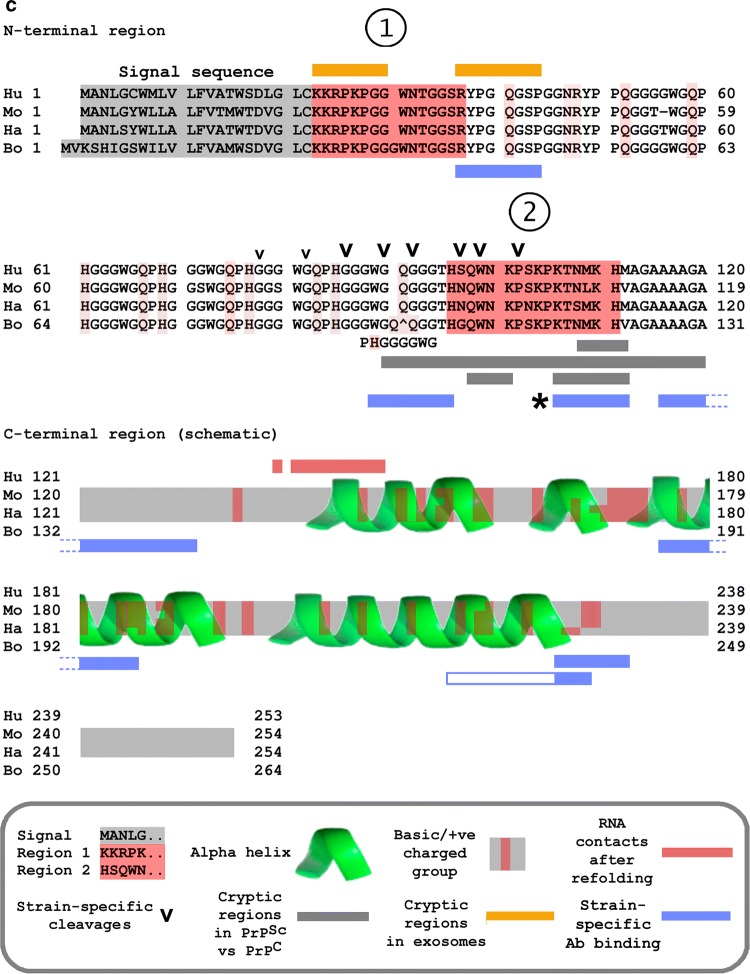
Fig. 1.
Two basic regions in the N-terminal segment of PrP and its immediate evolutionary precursors: TSE strain-specific structural differences. (A) The two polybasic regions in the intrinsically disordered N-terminus of human PrP, schematically depicted in complex with a nucleic acid (DNA for illustration), are shown fused to the globular/α-helical region of the protein established by NMR (PDB 1QM2 [236]). (B) Conservation of two basic regions in PrP and its evolutionary precursor (Shadoo/SPRN). PrP sequences (P) are Hu, human; Mo, mouse; Bo, bovine; Md, opossum (marsupial, Monodelphis domestica); Cs, mousebird (Colius striatus); Gg, chicken (Gallus gallus); Pm, viper (Protobothrops mucrosquamatus); Xl, African clawed frog (Xenopus laevis). The final three entries are prion protein 1 from zebrafish (DrP, Danio rerio), Shadoo/SPRN from zebrafish (DrS), and Shadoo/SPRN from whale shark (RtS, Rhincodon typus) which lack the C-terminal globular region and have alternative region 2 polybasic sequences (His-rich in DrP, and Arg-rich in DrS and RtS). Color code: red, highly basic (Lys/Arg); violet, basic (Asn/Gln/His); green, acidic (Asp/Glu); yellow, hydrophobic (Leu/Ile/Val/Met/Phe/Tyr). (C) TSE strains and the N-terminal region of PrP. Summary of strain-dependent cleavage sites, strain-specific antibody binding, occlusion of antibody binding, and RNA-dependent refolding of PrP. The figure shows the alignment of the N-termini of human (Hu), mouse (Mo), hamster (Ha), and bovine (Bo) PrPC sequences (the C-terminus is depicted schematically). Regions 1 and 2 are as in panels A and B. The exact details depend on the host species (and host genotype) as well as on the strain of the agent. Strain-specific cleavages (v) are sites where high-resolution mapping (e.g., mass spectrometry) demonstrates that the precise sites of PrP proteolytic processing differ significantly between infections with different strain types [119, 128, 129, 237]. Grey and brown horizontal bars show regions that are occluded (‘cryptic’) in PrPSc versus PrPC (grey) and that can also differ according to the strain of TSE [130–133], or that are occluded in exosomes from scrapie-infected cells (brown) [220]. Blue horizontal bars indicate epitopes for strain-specific antibody binding [238–240]. The epitope marked with an asterisk (*) is recognized by antibody 3F4 [241]; RNA binding to PrP in vitro occludes 3F4 binding (J-L.D, unpublished). Molecular dynamics simulations indicate that RNA bound to site 1 can lead to refolding of the polypeptide and dissolution of the first α-helical region; predicted new contacts after refolding are indicated by red bars; there may be further contacts within PrP [115]. ‘Basic’ amino acids indicated in the figure include not only K and R but also H, Q, and N, which contain positively charged groups with potential to interact with nucleic acid phosphates. The panel aims to highlight strain-specific differences and is not intended as a comprehensive survey