Table 2.
Structures and 1H-NMR characteristics (solvent: CD3CN:D2O 80:20) of standard monacolins and other compounds identified in this study.
| Compound | Structure | 1H-NMR 1 |
|---|---|---|
| δ (ppm) (Multiplicity 2, J (Hz), Number of Protons, Attribution) | ||
| Monacolin K lactone form (MK) | 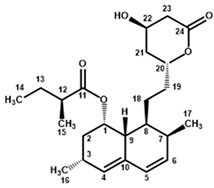 |
6.01 (d, J = 9.6, 1H, H-5), 5.84 (dd, J = 6.1, 9.6, 1H, H-6), 5.56 (app t, J = 2.8, 1H, H-4), 5.35 (q, J = 3.2, 1H, H-1), 4.59 (m, 1H, H-20), 4.25 (app quint, J = 3.9, 1H, H-22), 2.69 (Ad, J = 4.9, 17.6, 1H, H-23), 2.51 (Bdd, J = 1.7, 3.8, 17.6, 1H, H-23), 2.45 (m, 1H, H-3), 2.42 (m, 1H, H-7), 2.37 (m, 1H, H-9), 2.35 (m, 1H, H-12), 1.96 (m, 2H, H-2), 1.90 and 1.71 (two m, 2H, H-21), 1.81 and 1.37 (two m, 2H, H-19), 1.69 (m, 1H, H-8), 1.62 (app qd, J = 7.4, 13.6, 1H, H-13), 1.46 (m, 2H, H-13 and H-18), 1.36 (m, 1H, H-18), 1.08 (d, J = 6.9, 3H, H-15), 1.06 (d, J = 7.4, 3H, H-16), 0.89 (d, J = 6.9, 3H, H-17), 0.88 (t, J = 7.5, 3H, H-14) |
| Monacolin K hydroxyl acid form (MKA) | 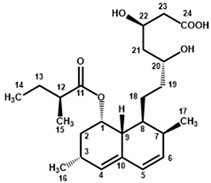 |
5.99 (d, J = 9.6, 1H, H-5), 5.83 (dd, J = 6.1, 9.6, 1H, H-6), 5.53 (app t, J = 2.8, 1H, H-4), 5.33 (q, J = 3.2, 1H, H-1), 4.05 (m, 1H, H-22), 3.63 (app hept, J = 4.1, 1H, H-20), 2.42 (m, 1H, H-3), 2.39 (m, 1H, H-7), 2.35 (m, 1H, H-9), 2.33 (m, 2H, H-12 and H-23), 2.16 (dd, J = 8.7, 15.2, 1H, H-23), 1.93 (m, 2H, H-2), 1.63 (m, 1H, H-8), 1.59 and 1.45 (two m, 2H, H-13), 1.57 and 1.51 (two m, 2H, H-21), 1.53 and 1.15 (two m, 2H, H-19), 1.32 (m, 2H, H-18), 1.08 (d, J = 6.9, 3H, H-15), 1.05 (d, J = 7.4, 3H, H-16), 0.87 (d, J = 6.9, 3H, H-17), 0.86 (t, J = 7.4, 3H, H-14) |
| Compactin (CP) = Mevastatin | 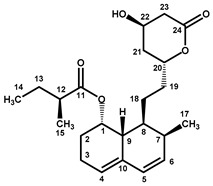 |
6.00 (d, J = 9.7, 1H, H-5), 5.79 (dd, J = 6.0, 9.7, 1H, H-6), 5.57 (m, 1H, H-4), 5.30 (m, 1H, H-1), 4.60 (m, 1H, H-20), 4.25 (app quint, J = 3.9, 1H, H-22), 2.69 (Ad, J = 4.8, 17.6, 1H, H-23), 2.51 (Bdd, J = 1.7, 3.6, 17.6, 1H, H-23), 2.42 (m, 2H, H-7 and H-9), 2.38 (m, 1H, H-12), 2.15 (m, 2H, H-3), 2.08 (m, 1H, H-2), 1.90 (m, 1H, H-21), 1.81 and 1.37 (two m, 2H, H-19), 1.72 (m, 2H, H-2 and H-21), 1.68 (m, 1H, H-8), 1.62 (app qd, J = 7.6, 13.6, 1H, H-13), 1.48 and 1.39 (two m, 2H, H-18), 1.46 (m, 1H, H-13), 1.11 (d, J = 7.0, 3H, H-15), 0.90 (d, J = 7.0, 3H, H-17), 0.89 (t, J = 7.5, 3H, H-14) |
| Dehydromonacolin K (DeMK) |
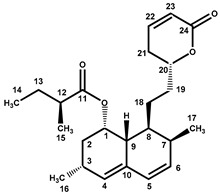 |
7.03 (ddd, J = 2.5, 6.0, 9.8, 1H, H-22), 6.02 (d, J = 9.6, 1H, H-5), 5.97 (ddd, J = 1.0, 2.7, 9.8, 1H, H-23), 5.84 (dd, J = 6.1, 9.6, 1H, H-6), 5.56 (app t, J = 2.8, 1H, H-4), 5.33 (q, J = 3.2, 1H, H-1), 4.42 (m, 1H, H-20), 2.44 (m, 1H, H-3), 2.43 (m, 1H, H-7), 2.42 and 2.30 (two m, 2H, H-21), 2.36 (m, 1H, H-9), 2.35 (m, 1H, H-12), 1.96 (m, 2H, H-2), 1.90 and 1.41 (two m, 2H, H-19), 1.70 (m, 1H, H-8), 1.62 (app qd, J = 7.5, 13.8, 1H, H-13), 1.46 (m, 1H, H-13), 1.45 and 1.39 (two m, 2H, H-18), 1.09 (d, J = 6.9, 3H, H-15), 1.07 (d, J = 7.4, 3H, H-16), 0.89 (d, J = 6.9, 3H, H-17), 0.88 (t, J = 7.5, 3H, H-14) |
| Dihydromonacolin K (DiMK) |
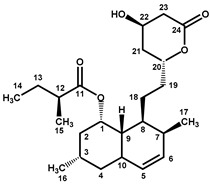 |
5.69 (ddd, J = 2.7, 5.0, 9.8, 1H, H-6), 5.42 (d, J = 9.8, 1H, H-5), 5.15 (q, J = 2.7, 1H, H-1), 4.58 (m, 1H, H-20), 4.25 (app quint, J = 3.8, 1H, H-22), 2.68 (Ad, J = 4.9, 17.7, 1H, H-23), 2.50 (Bdd, J = 1.7, 3.7, 17.7, 1H, H-23), 2.46 (m, 1H, H-10), 2.39 (m, 1H, H-12), 2.34 (m, 1H, H-7), 2.05 (m, 1H, H-3), 1.88 and 1.72 (two m, 2H, H-21), 1.81 and 1.37 (two m, 2H, H-19), 1.80 (m, 2H, H-2), 1.66 (m, 1H, H-8), 1.64 (m, 1H, H-13), 1.61 and 1.35 (two m, 2H, H-4), 1.49 (app qd, J = 7.4, 13.6, 1H, H-13), 1.35 and 1.30 (two m, 2H, H-18), 1.12 (d, J = 7.0, 3H, H-15), 1.11 (d, J = 7.6, 3H, H-16), 0.90 (t, J = 7.5, 3H, H-14), 0.86 (d, J = 7.0, 3H, H-17) |
| Monascin 3 | 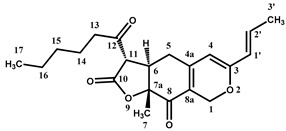 |
6.50 (qd, J = 7.0, 15.5, 1H, H-2′), 6.05 (qd, J = 1.7, 15.5, 1H, H-1′), 5.51 (s, 1H, H-4), 4.97 (At, J = 0.9, 12.7, 1H, H-1), 4.72 (Bt, J = 1.4, 12.7, 1H, H-1), 4.14 (d, J = 13.3, 1H, H-11), 3.15 (m, 1H, H-6), 2.88 (At, J = 7.3, 18.2, 1H, H-13), 2.67 (Bt, J = 7.3, 18.2, 1H, H-13), 2.63 (app br d, J = 7.7, 2H, H-5), 1.87 (dd, J = 1.7, 7.0, 3H, H-3′), 1.59 (quint, J = 7.3, 2H, H-14), 1.43 (s, 3H, H-7), 1.32 (m, 4H, H-15, H-16), 0.91 (t, J = 7.2, 3H, H-17) |
| Citrinin 4 | 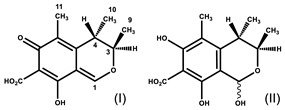 |
8.45 (br s, H-1 (I)), 5.94 (s, H-1 (II major), 5.90 (br s, H-1 (II minor)), 4.92 (br signal, H-3 (I)), 4.09 (quint, J = 6.6, H-3 (II major and minor)), 3.15 (br signal, H-4 (I)), 2.76 (br signal, H-4 (II minor)), 2.67 (quint, J = 6.8, H-4 (II major)), 2.03 (s, H-11 *), 2.01 (s, H-11 *), 2.00 (s, H-11 *), 1.30 (d, J = 6.7, H-9 *), 1.27 (d, J = 6.4, H-9 *), 1.20 (d, J = 6.9, H-10 *), 1.17 (very br signal, H-10 *) |
| Piperine | 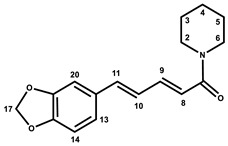 |
7.28 (ddd, J = 2.7, 7.3, 14.7, 1H, H-9), 7.11 (d, J = 1.6, 1H, H-20), 6.99 (dd, J = 1.7, 8.1, 1H, H-13), 6.87 (d, J = 8.0, 1H, H-14), 6.83 (m, 2H, H-10, H-11), 6.60 (d, J = 14.7, 1H, H-8), 6.00 (s, 2H, H-17), 3.55 (m, 4H, H-2, H-6), 1.65 (m, 2H, H-4), 1.56 (m, 4H, H-3, H-5) |
| Carnitine | 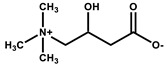 |
4.56 (br q, J = 7.1, 1H, CH-OH), 3.39 (m, 2H, CH2-N+), 3.17 (s, 9H, (CH3)3-N+), 2.58 (m, 2H, CH2-COO−) |
| Chlorogenic acid | 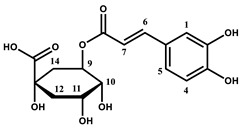 |
7.61 (d, J = 15.9, 1H, H-6), 7.16 (d, J = 2.0, 1H, H-1), 7.06 (dd, J = 2.0, 8.2, 1H, H-5), 6.88 (d, J = 8.2, 1H, H-4), 6.34 (d, J = 15.9, 1H, H-7), 5.28 (ddd, J = 4.6, 9.4, 10.5, 1H, H-9), 4.18 (app q, J = 3.5, 1H, H-10), 3.75 (dd, J = 3.2, 9.4, 1H, H-11), 2.26–1.97 (m, 4H, H-12, H-14) |
| Saturated fatty acids | 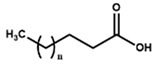 |
2.20 (t, J = 7.5, 2H, CH2-COOH), 1.59 (m, 2H, CH2-CH2-COOH), 1.30 (m, 2nH, (CH2)n), 0.90 (t, J = 6.3, 3H, CH3) |
| α/β Glucose | 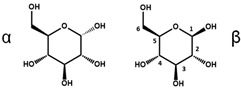 |
5.14 (d, J = 3.7, 1H, H-1α), 4.52 (d, J = 7.9, 1H, H-1β), 3.85-3.60 (m, 7H, H-3α, H-4α/β, H-6α/β), 3.45–3.28 (m, 4H, H-2α, H-3β, H-5α/β), 3.15 (t, J = 8.5, 1H, H-2β) |
| Glycerol | 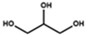 |
3.66 (tt, J = 4.6, 6.3, 1H, CH), 3.55 (Ad, J = 4.6, 11.5, 2H, CH2), 3.48 (Bd, J = 6.2, 11.5, CH2) |
| Isopropyl alcohol | 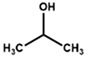 |
3.93 (hept, J = 6.1, 1H, CH), 1.14 (d, J = 6.1, 6H, CH3) |
| Linoleic acid 5 | 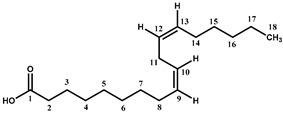 |
5.36 (m, 4H, H-9, H-10, H-12, H-13), 2.78 (t, J = 6.7, 2H, H-11), 2.27 (t, J = 7.5, 2H, H-2), 2.06 (m, 4H, H-8, H-14), 1.56 (m, 2H, H-3), 1.40-1.27 (m, 14H, H-4, H-5, H-6, H-7, H-15, H-16, H-17), 0.89 (t, J = 6.8, 3H, H-18) |
| Sorbitol | 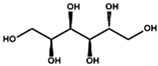 |
3.82–3.54 (m, 8H) |
| Vitamin B3 (niacinamide form) |
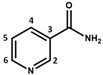 |
8.99 (d, J = 2.3, 1H, H-2), 8.72 (dd, J = 4.9, 1.6, 1H, H-6), 8.25 (td, J = 8.0, 1.9, 1H, H-4), 7.56 (dd, J = 8.0, 4.9, 1H, H-5) |
| Vitamin C | 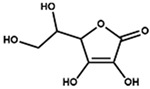 |
4.76 (d, X part of an ABMX system, JXM = 2.0, 1H, CH-O-), 3.94 (td, M part of an ABMX system, JMX = 1.9, JMA = JMB = 6.6, 1H, CH-OH), 3.66 (AB part of an ABMX system, JAB = 11.3, JAM = JBM = 6.6, 2H, CH2-OH) |
1 The assignments of 1H-NMR signals of monacolins, citrinin, and monascin were in agreement with the literature data even if the solvents were different: CDCl3 for MK, MKA, CP, DeMK, DiMK and citrinin [1,20,21,22,23,24], DMSO-d6 for monascin [25] and D2O for citrinin [22]. 2 d: doublet, dd: doublet of doublet, ddd: doublet of doublet of doublet, t: triplet, td: doublet of triplet, tt: triplet of triplet, q: quadruplet, qd: doublet of quadruplet, quint: quintuplet, hept: heptuplet, m: multiplet, A: part A of an AB system, B: part B of an AB system, app: apparent, br: broad. 3 H11 is exchanged with D and its resonance disappears rapidly with time as well as its coupling with H6 whose multiplet becomes a triplet (J = 7.9 Hz). 4 The attributions were done according to the literature [22]. Citrinin exists as the quinone methide (I) in organic NMR solvents but as a diastereoisomeric mixture of hydrates (II) in aqueous solution at physiological pH, one diastereoisomer being major and the other minor. As the solvent used in the present study is a CD3CN:D2O (80:20) mixture, the two forms (I) and (II) are observed. * From the literature data, it was not possible to assign the resonances of H9, H10 and H11 to a specific form. 5 As a model of non-conjugated unsaturated fatty acids.
