Figure 4.
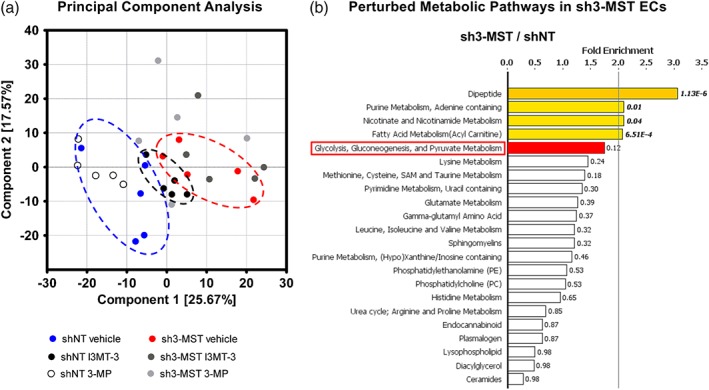
3‐MST down‐regulation alters EC metabolism. Global metabolomic profiling was performed on shNT and sh3‐MST #2 ECs with three treatment groups each—vehicle, I3MT‐3 (100 μM), and 3‐MP (300 μM)—to determine the concentration of 669 metabolic compounds in each sample. (a) Principal component analysis (PCA) showed primary separation of samples related to 3‐MST silencing from shNT samples, and lesser extent samples related to 3‐MST inhibition. PCA analysis also indicates that (a) primarily, the enzymic activity of 3‐MST regulates the EC metabolome and (b) 3‐MST pharmacological inhibition partly reproduces the overall metabolic effects of 3‐MST silencing. (b) 3‐MST silencing induced alterations (increased or reduced number of metabolites) in many pathways. The five metabolic pathways most affected by 3‐MST silencing were shown to be dipeptide, purine, nicotinamide, fatty acid, and glucose metabolism. Sulfur metabolism ranked seventh on this list. Several sub‐pathways of lipid, amino acid, and nucleotide metabolism were also seen. Fold enrichment (FE) values were calculated by the following equation using MetaboLync online software: FE = (# of significant metabolites in pathway [k] /total # of detected metabolites in pathway[m] / (total # of significant metabolites[n] /total # of detected metabolites [N]); in short: FE = (k/m)/(n/N). A pathway FE value >1 indicated that the pathway contained more experimentally regulated compounds relative to the overall study, suggesting that the pathway may be a target of interest of the experimental perturbation. P values displayed next to each FE bar were calculated using a two‐way ANOVA. FE bars representing the most perturbed pathways are shaded yellow (FE > 2) or red (1.5 < FE < 2). Only major pathways with over 10 metabolites were displayed. All six groups consisted of data of five independent experiments (n = 5).
