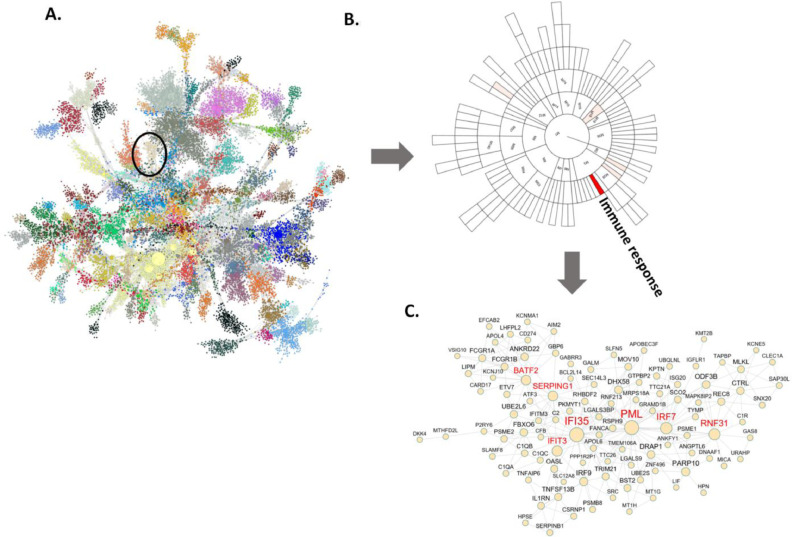
Fig. 3.
Gene co-expression network analysis of the gene expression data from the blood samples of the AD and normal control subjects identifies an immune enriched gene module associated with the DTI features. (A) The global gene co-expression network is comprised of highly co-expressed modules which are highlighted in different colors. Co-expression network is constructed based on the significant correlations among gene expression profiles and then used to identify clusters or modules of highly interconnected genes. Associations between the DTI features and the module eigenvectors (the principal components of modules) are calculated to rank order the gene modules. The most significant module (M30) is highlighted in a black circle. (B) A sunburst plot showing module hierarchy and the associations between the modules and the DTI features across all the brain regions. Each block represents a module and the color intensity of a block indicates the correlation strength between the corresponding module eigenvector and the DTI features (FA, PLA, LIN, MD and RD) with the solid red color standing for the strongest correlation (M30) and the white color for no significant correlation. The M30 was ranked the top based on the summary of all the significant correlations with 9 DTI features. (C) The network plot of the module (M30) that is most significantly associated with the DTI features. The nodes with red labels are the hubs in the network. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)