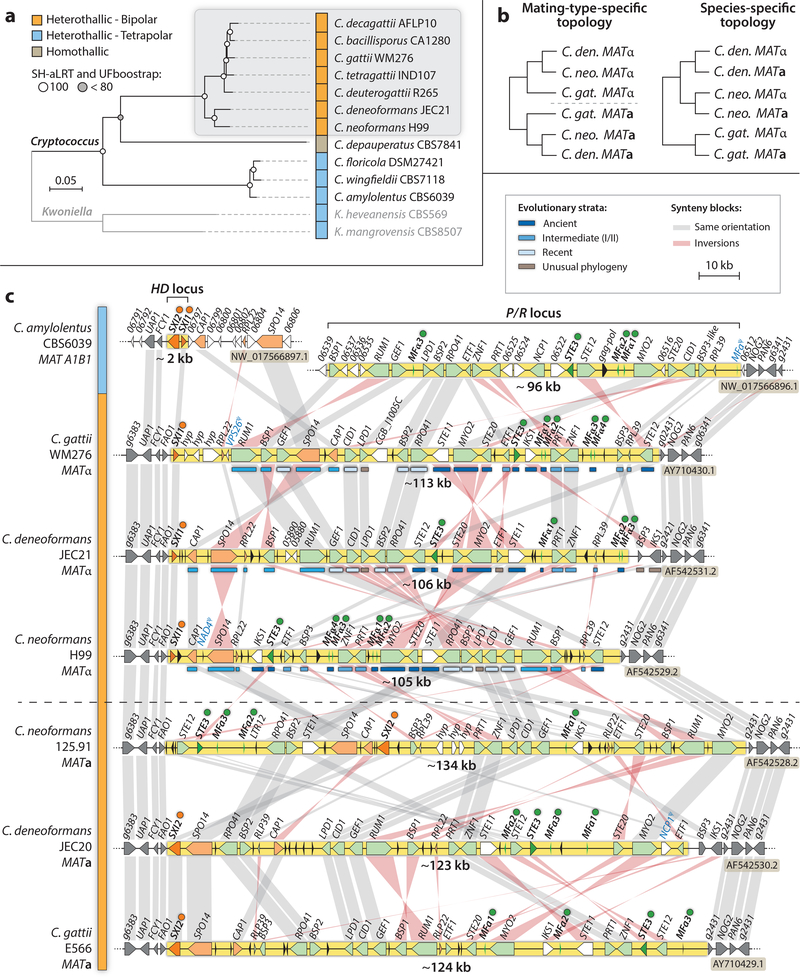
Figure 3. Cryptococcus phylogeny and comparative analyses of MAT alleles from pathogenic and closely related non-pathogenic Cryptococcus species.
(a) Seven species are currently recognized among the pathogenic C. neoformans/C. gattii species complex (shadowed in grey), all of which have linked P/R and HD mating-type loci (bipolar). Conversely, C. amylolentus, C. floricola, C. wingfieldii, and the two Kwoniella species (outgroup) are tetrapolar, with mating-type genes at two genetic loci on different chromosomes representing the ancestral configuration of the MAT loci. The tree was inferred from 734 single copy orthologs (149,332 informative amino acid sites), using maximum likelihood and the LG+F+R4 model of amino acid substitution in IQ-TREE. Branch support was assessed by ultrafast bootstrap (UFboot) and SH-aLRT methods. Scale bar indicates the number of substitutions per site. (b) Example of tree topologies observed for genes anciently (left) or recently (right) recruited to MAT, representing different evolutionary strata. (c) The MAT alleles of the pathogenic species are highly rearranged between species and mating types and result from linkage between P/R and HD gene clusters located on different chromosomes in C. amylolentus. The nonrecombining MAT region is depicted in yellow (with the number beneath indicating the approximate size), and is bordered by ~10 kb of common flanking regions depicted on each side (4 genes in dark grey), except in C. deneoformans where the IKS1 and BSP3 genes were evicted from MAT and fixed in the flanking region by inversion and recombination. Common genes found in the HD and P/R regions of C. amylolentus are colored in orange and green, respectively, in the extant MAT locus of the pathogenic species. Genes encoding homeodomain transcription factors (SXI1 and SXI2), pheromones (MF), and pheromone receptors (STE3) are bulleted. Blue bars below the genes indicate the evolutionary strata class they belong to as shown in the key. Genes in both the “Ancient” and the “Intermediate” strata classes have mating-type specific phylogenies (they entered MAT prior to speciation), while genes in the “Recent” strata class display species-specific phylogenies (they began diverging only after speciation). Genes with unusual phylogenies represent cases where the gene underwent gene conversion in one or the other lineage, thereby fixing one of the two alleles in both mating types and with concomitant loss of the alternative allele. Grey and pink bars connect orthologs with the same or inverted orientation, respectively. Genes depicted in white are not conserved in all species. Pseudogenes are labeled in blue, and black arrows represent transposable elements or their remnants.