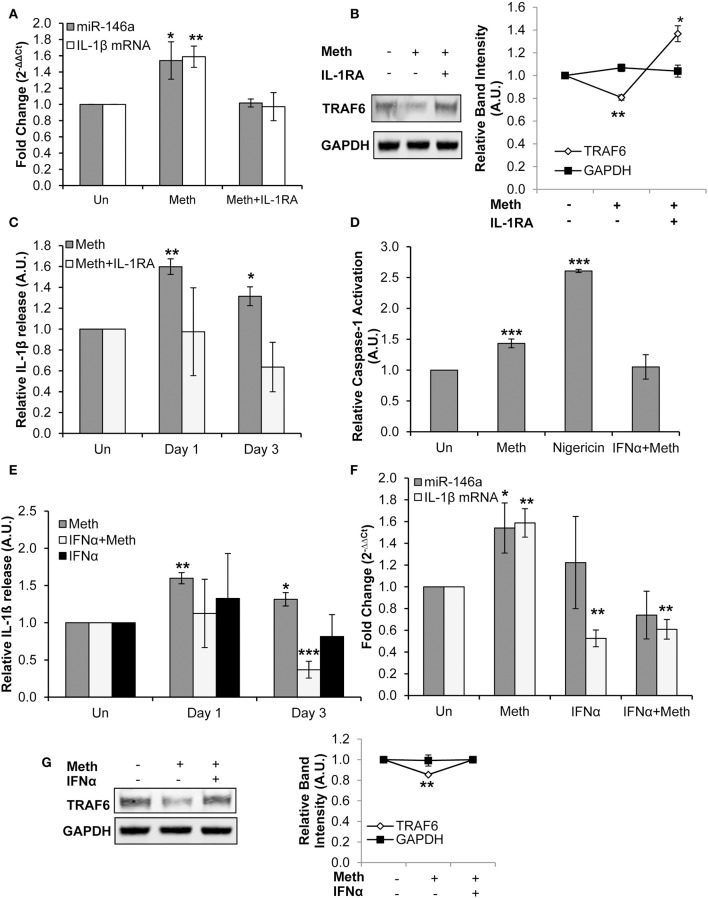
Figure 4.
Meth increases miR-146a and IL-1β mRNA expression via IL-1 signaling. (A) CD4+ T-cells treated with or without Meth for 3 days and IL-1RA were analyzed for miR-146a and IL-1β mRNA expression by RT-qPCR. Fold change was calculated by normalizing Meth treated and Meth+IL-1RA treated cells to untreated controls. Data represent the mean ± SD of 3 independent experiments, and p-values were calculated relative to untreated controls (*p < 0.05, **p < 0.01). (B) Protein extracts from cells treated for 3 days with or without Meth and IL-1RA were analyzed for TRAF6 by Western Blotting. GAPDH was used as a loading control. Relative band intensity was calculated using ImageJ software, and p-values were calculated relative to untreated controls (*p < 0.05, **p < 0.01). (C) Culture supernatants were harvested after 3 days of treatment and analyzed for IL-1β by ELISA. Relative expression was calculated by normalizing Meth and Meth+IL-1RA treated samples to untreated controls. Data represent the mean ± SD of 3 independent experiments, and p-values were calculated relativeto untreated controls (*p ≤ 0.05, **p ≤ 0.01). (D) CD4+ T-cells were untreated, treated with Meth, treated with Nigericin, or treated with IFNα and Meth for 24 h. Caspase-1 Activation was measured using fluorescent labeling with FAM-FLICA, and analyzed by Flow Cytometry. Data represent the mean ± SD of 3 independent experiments, and p values were calculated relative to untreated controls (***p < 0.001). (E) CD4+ T-cells were untreated, treated with Meth, or treated with IFNα and Meth, daily for 3 days. Culture supernatants were analyzed for IL-1β expression by ELISA. Relative expression was calculated by normalizing Meth treated samples to untreated controls. Data represent the mean ± SD of 3 independent experiments, and p-values were calculated relative to untreated controls (*p < 0.05, **p < 0.01, ***p < 0.001). (F) Cells were untreated, treated with Meth, or treated with IFNα and Meth, daily for 3 days. miR-146a and IL-1β mRNA expression were determined by RT-qPCR. Fold change was calculated by normalizing Meth treated and Meth+IFNα treated cells to untreated controls. Data represent the mean ± SD of 3 independent experiments, and p-values were calculated relative to untreated controls (*p < 0.05, **p < 0.01). (G) Cells were untreated, treated with Meth, or treated with IFNα and Meth, daily for 3 days. Protein extracts were analyzed for TRAF6 by Western Blot. GAPDH was used as a loading control. Relative band intensity was calculated using ImageJ software, and p-values were calculated relative to untreated controls (**p < 0.01).