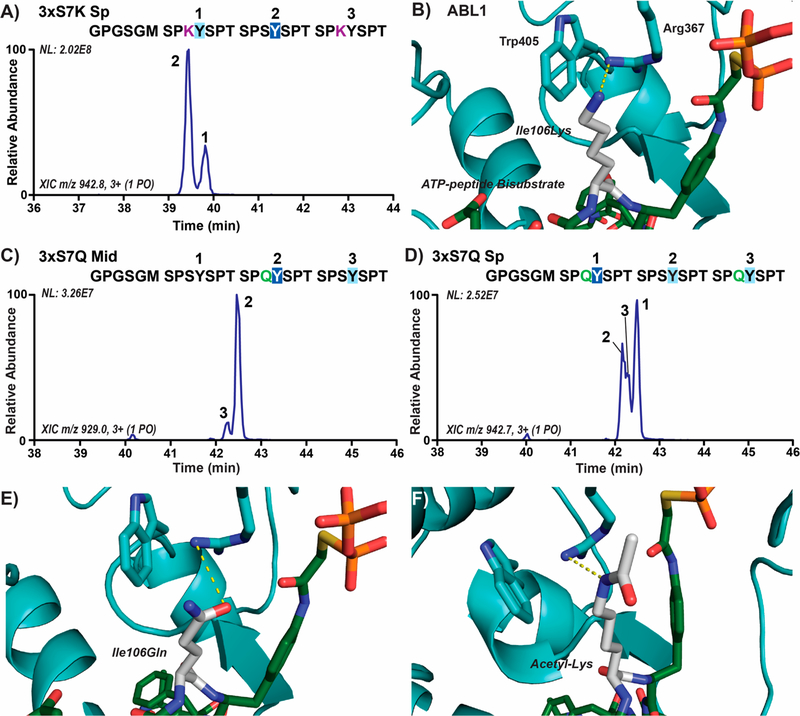
Figure 6.
Lysine and an acetylated lysine mimic (glutamine) in the 7th CTD position alter ABL1 preference for Tyr1 sites. (A) Extracted chromatograms of singly phosphorylated species in three-repeat CTD substrates with Ser7Lys mutations (3xS7K Sp) treated with ABL1. Phosphorylation sites were localized using UVPD-MS and are indicated on the chromatogram by repeat number UVPD-MS. The GPGSGM amino acid sequence at the N-terminus is retained after 3C proteolysis of the 6xHis-GST tags prior to mass spectrometry. The sites of major and minor phosphorylation were indicated by dark or light blue boxes, respectively, and the lysine mutations are shown in purple. (B) Structure of ABL1 (light blue) bound to the ATP-peptide bisubstrate analog of SRC (green) with Ile106 modeled as a lysine (white). (C,D) Extracted chromatograms of singly phosphorylated species in three-repeat CTD substrates with acetyl-lysine mimicking Ser7Gln mutations (3xS7Q Mid/Sp) treated with ABL1. (E,F) Structure of ABL1 (light blue) bound to the ATP-peptide bisubstrate analog of SRC (green) with Ile106 modeled as either acetyl-lysine mimicking glutamine or acetyl-lysine (white).