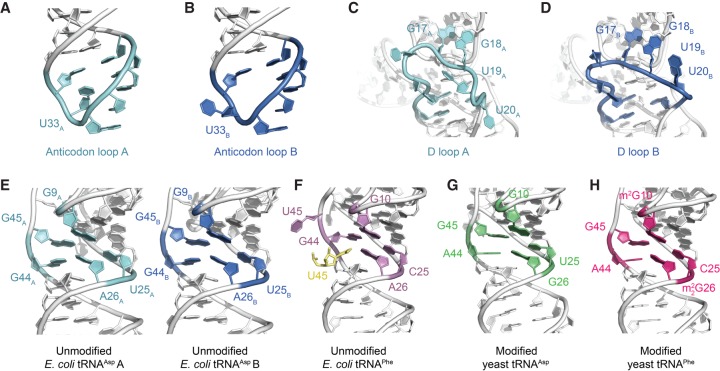
FIGURE 2.
Comparative structural analyses between the crystal structure of E. coli tRNAAsp with other free tRNA molecules. (A,B) Conformational differences in the anticodon loop of the two molecules of E. coli tRNAAsp that comprise the crystallographic asymmetric unit. (C,D) Conformational differences in the D loop of the two molecules of E. coli tRNAAsp that comprise the crystallographic asymmetric unit. (E–H) The core region of the two molecules of unmodified E. coli tRNAAsp that comprise the crystallographic asymmetric unit is very similar in conformation to the core region of both modified yeast tRNAAsp (PDB ID: 1VTQ) (Comarmond et al. 1986) and yeast tRNAPhe (PDB ID: 1EHZ) (Shi and Moore 2000), whereas the core region of an unmodified E. coli tRNAPhe (PDB ID: 3L0U) (Byrne et al. 2010) is comparatively different (m2G and m22G represent 2-methylguanosine and N2,N2-dimethylguanosine, respectively). In the latter structure, the extruded U45 forms a crystal lattice contact and is replaced by U45 from a symmetry related molecule (yellow).