Fig. 5.10.
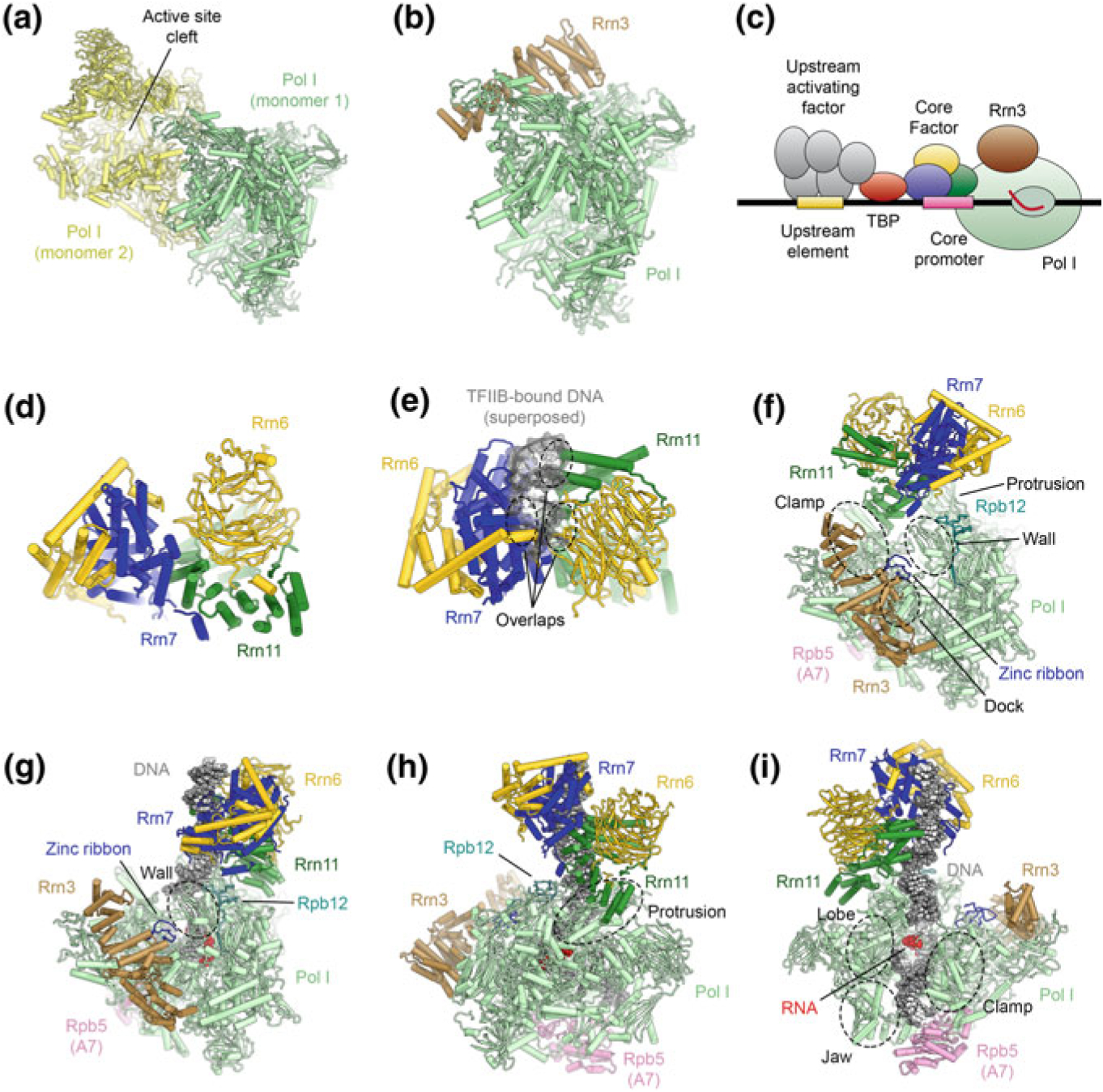
The architecture of the RNA polymerase I PIC and ITC. a Structure of the Pol I-dimer (Engel et al. 2013), with a widened active site cleft, as observed in Pol I crystals. b Structure of Pol I bound with Rrn3 (Engel et al. 2016), which blocks the Pol I dimerization interface. c Schematic of the Pol I core promoter and general transcription factors in yeast. d Structure of yeast core factor (CF) (Engel et al. 2017). e Structure of yeast core factor shown with DNA superposed from a TFIIB-DNA structure (Tsai and Sigler 2000). The superposed DNA overlaps with CF, indicating that Rrn7 in CF cannot bind to DNA by the same mechanism as TFIIB (also see (Fig. 5.15). f Structure of the Pol I-Rrn3-CF complex (Engel et al. 2017). Regions of Pol I that are in contact with CF are indicated. g–i Depiction of the Pol I-ITC, with bound DNA, Rrn3, and CF (Engel et al. 2017; Han et al. 2017). Contacts differ from the DNA-free complex (shown in f) and corresponding regions on Pol I are indicated. Downstream promoter DNA is stabilized by contacts from the Pol I clamp, lobe, and jaw domains and Rpb5
