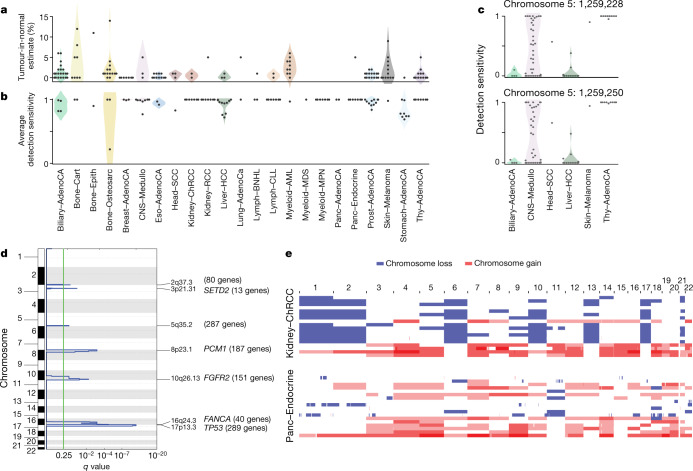
Fig. 3. Analysis of patients with no detected driver mutations.
a, Individual estimates of the percentage of tumour-in-normal contamination across patients with no driver mutations in PCAWG (n = 181). No data were available for myelodysplastic syndromes and acute myeloid leukaemia. Points represent estimates for individual patients, and the coloured areas are estimated density distributions (violin plots). Abbreviations of the tumour types are defined in Extended Data Table 1. b, Average detection sensitivity by tumour type for tumours without known drivers (n = 181). Each dot represents a given sample and is the average sensitivity of detecting clonal substitutions across the genome, taking into account purity and ploidy. Coloured areas are estimated density distributions, shown for cohorts with at least five cases. c, Detection sensitivity for TERT promoter hotspots in tumour types in which TERT is frequently mutated. Coloured areas are estimated density distributions. d, Significant copy-number losses identified by two-sided hypothesis testing using GISTIC2.0, corrected for multiple-hypothesis testing. Numbers in parentheses indicate the number of genes in significant regions when analysing medulloblastomas without known drivers (n = 42). Significant regions with known cancer-associated genes are labelled with the representative cancer-associated gene. e, Aneuploidy in chromophobe renal cell carcinomas and pancreatic neuroendocrine tumours without known drivers. Patients are ordered on the y axis by tumour type and then by presence of whole-genome duplication (bottom) or not (top).