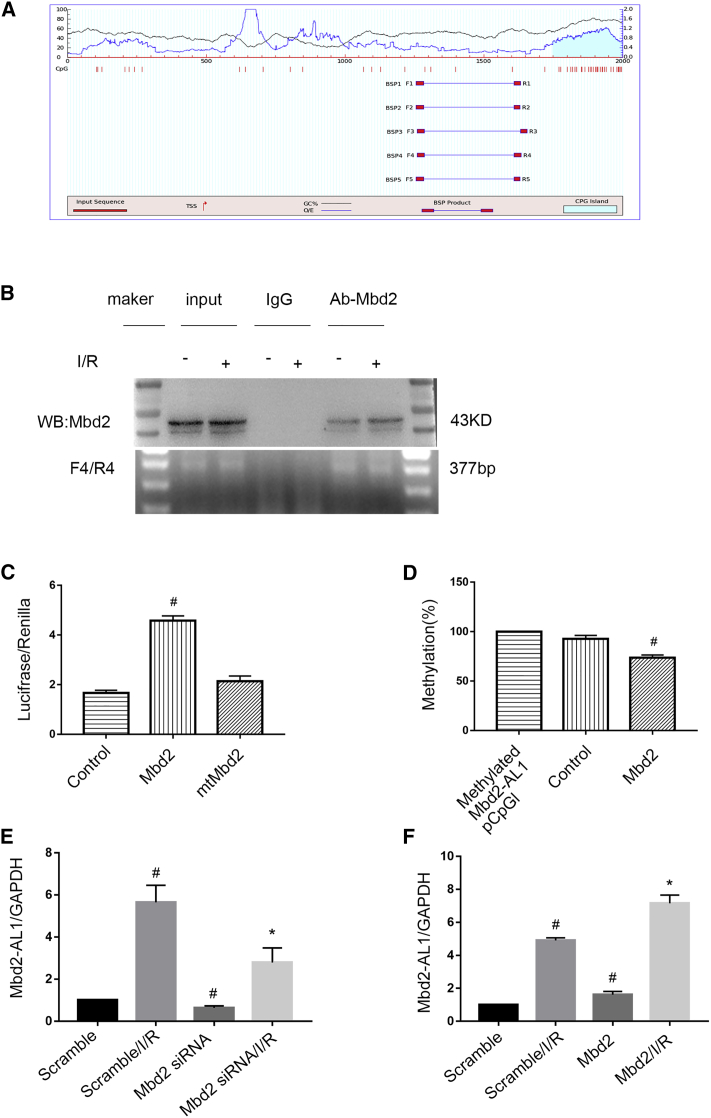
Figure 4.
Mbd2 Suppresses the Methylation of the lncRNA Mbd2-AL1 Promoter and Activates Its Demethylation
(A) The CpG island of the Mbd2-AL1 promoter was predicted, and five primer pairs were designed by the software MethPrimer 2.0. (B) ChIP assays were performed with chromatin materials, isolated from RGCs, treated with I/R, and precipitated with Mbd2, IgG, or without antibody (input) and used as a template for PCR detection of potential Mbd2 binding site 4 (mBS4). (C) Relative luciferase activity in RGCs. Cotransfection of Mbd2 plasmid, mtMbd2 plasmid, or control with the pCpGfree-basic luciferase reporter plasmid containing the promoter region of Mbd2-AL1. The data were expressed as mean ± SEM of five independent experiments. #p < 0.05 versus mtMbd2. (D) The percentage of CpG-DNA methylation of the Mbd2-AL1 promoter. Cotransfection of the Mbd2 plasmid or control with the pCpGfree-basic luciferase reporter plasmid containing the methylated promoter region of Mbd2-AL1. The data were expressed as mean ± SEM of five independent experiments. #p < 0.05 versus the control group. (E and F) Quantitative real-time PCR of the expression of lncRNA Mbd2-AL1 in RGCs. (E) Mbd2 siRNA suppressed the expression of Mbd2-AL1. (F) The expression of Mbd2-AL1 in RGCs was upregulaed after transfection of exogenous Mbd2 plasmid. The data were expressed as mean ± SEM of five independent experiments. #p < 0.05 versus the scramble group; *p < 0.05 versus the scramble/I/R group.