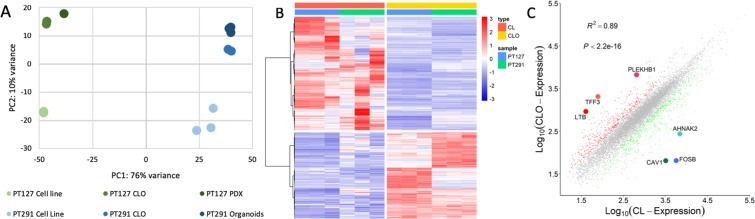
Figure 5.
(A) Principal component analysis was performed to compare the expression profiles of PT127 Cell line, PT127 CLO, PT127 PDX, PT291 cell line, PT291 CLO and PT291 organoids (B) Clustering of all differentially expressed genes (fold change >1, p-adj = 0.001) in CLOs and cell lines (PT127 and PT291) (C) Scatterplot – grouped PT291 and PT127 CLO vs CL, genes with the biggest log fold-change highlighted. FOSB (log fold change −6.496, p-adj 6.65E-50), CAV1 (log fold change −5.7652 p-adj 4.89E-75), AHNAK2 (log fold change −4.7934931, p-adj 9.25E-20), TFF3 (log fold change 4.8678, p-adj 5.86E-32), PLEKHB1 (log fold change 3.803, p-adj 2.59E-13), LTB (log fold change 3.633, p-adj 2.32E-11).