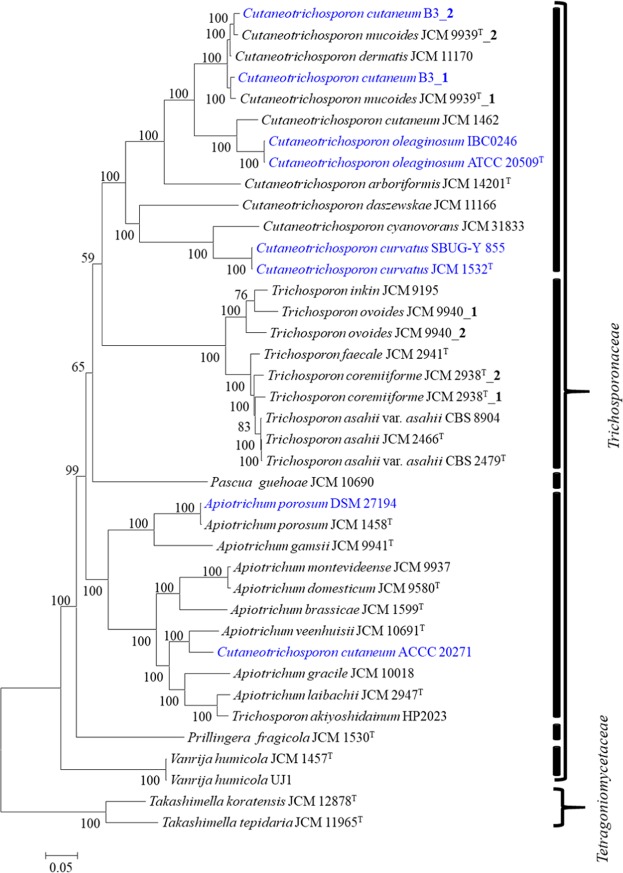
Figure 1.
Phylogenomic analysis of members of the family Trichosporonaceae. The maximum likelihood (ML) tree was inferred from the concatenated protein alignment (223,082 amino acids) of 405 proteins present in single copies among the haploid genomes and only in duplicate copies in the hybrid genomes. The phylogeny was generated using IQ-TREE version 1.6.7 based on the LG + F + R10 model. The ML was generated with confidence values based on 1,000 bootstrap replicates. The documented oil accumulating members of the family are indicated in blue fonts. The labels ‘_1’ and ‘_2’ indicate the two sets of single copy orthologs (SCOs) in the hybrid genomes, where the letter shows higher amino acid similarity to the closest haploid genome.