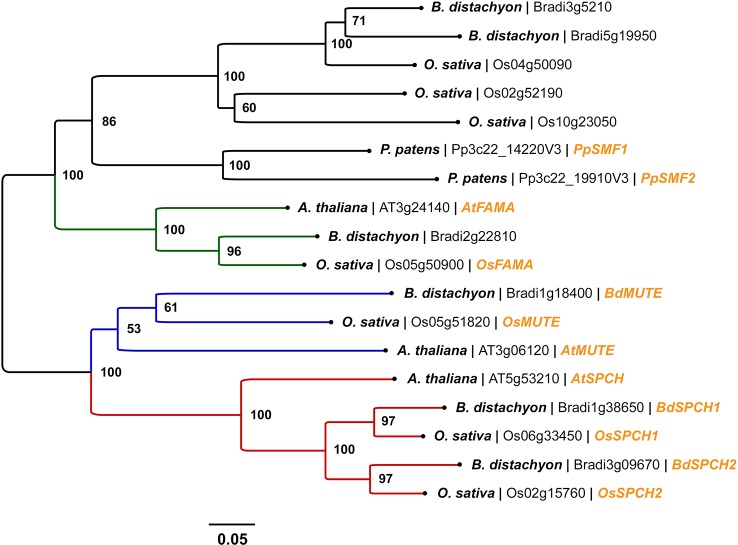
Figure 3.
Phylogenetic analysis of SPEECHLESS (SPCH), MUTE, and FAMA peptide sequences in Arabidopsis (Arabidopsis thaliana), rice (Oryza sativa), and Brachypodium (Brachypodium distachyon). Peptide sequences were obtained via BLAST searches of the rice and Brachypodium genomes using Phytozome v12. All peptide sequences with expect values (E-values) < 1 x 10-30 against AtSPCH, AtMUTE, and AtFAMA were used in the analysis. Stomatal bHLH peptide sequences PpSMF1 and PpSMF2 from the non-vascular moss Physcomitrella patens were included for evolutionary context; obtained from Chater et al. (2017). A total of 18 peptide sequences were involved in the phylogenetic analysis. Peptide sequences were aligned using the MUSCLE algorithm (Edgar, 2004). The phylogenetic tree was constructed using the neighbor-joining method (Saitou and Nei, 1987). The optimal tree with the sum of branch length = 4.05347046 is shown. A bootstrap test was performed (1,000 replicates) and the percentage of replicate trees that clustered taxa into the conformation displayed is shown adjacent to the nodes. Evolutionary distances between peptide sequences (in units of amino acid substitutions per site) were calculated using the Poisson correction method (Zuckerkandl and Pauling, 1965). The tree is drawn to scale, with branch lengths proportionate to the evolutionary distances used to infer the tree. All evolutionary analyses were performed in MEGA6. From left to right, species name, gene accession number and gene name (if characterised) are labeled at the branch tips. The clades comprising SPCH, MUTE, and FAMA orthologues are coloured red, blue and green respectively.