Figure 3.
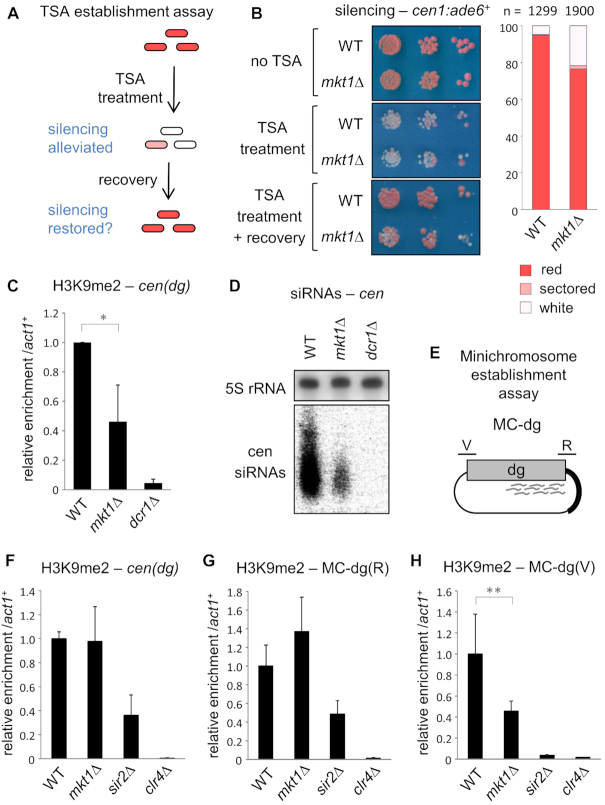
Mkt1 is required for efficient establishment of pericentromeric heterochromatin. (A) Schematic representation of the TSA heterochromatin re-establishment assay. Cells carrying the cen1:ade6+ reporter are treated with the HDAC inhibitor trichostatin A (TSA) which causes loss of H3K9me and silencing at cen1:ade6+, resulting in pink/white colonies on low adenine media. Following a period of recovery from TSA treatment, wild-type cells re-establish silencing resulting in red colonies; continued appearance of pink/white colonies indicates a defect in heterochromatin establishment. (B) Assay for silencing of cen1:ade6+. Cells were plated on low adenine media prior to TSA treatment (no TSA), after growth for 10 generations in medium containing 35 μg/ml TSA (TSA treatment), and after growth for a further 10 generations in the absence of TSA (TSA treatment + recovery). The chart shows the frequency of red, white and sectored colonies following TSA treatment and recovery. (C) ChIP-qPCR analysis of H3K9me2 levels at cen(dg) relative to act1+, normalised to wild-type, following TSA treatment and recovery. (D) Northern analysis of centromeric siRNAs following TSA treatment and recovery (5S rRNA is a loading control). (E) Schematic representation of the MC-dg plasmid used for the minichromosome establishment assay. The plasmid carries centromere central core sequence plus 5.6 kb of dg outer repeat sequence. The locations of primers used to monitor H3K9 methylation over the siRNA-rich region (R) and siRNA void region (V) are indicated. (F-H) In cells transformed with MC-dg, ChIP-qPCR analysis of H3K9me2 levels at cen(dg), MC-dg(R) and MC-dg(V), relative to act1+, normalised to wild-type. In each case data are averages of three biological replicates and error bars represent one SD; *P < 0.05, **P < 0.01.