Figure 3.
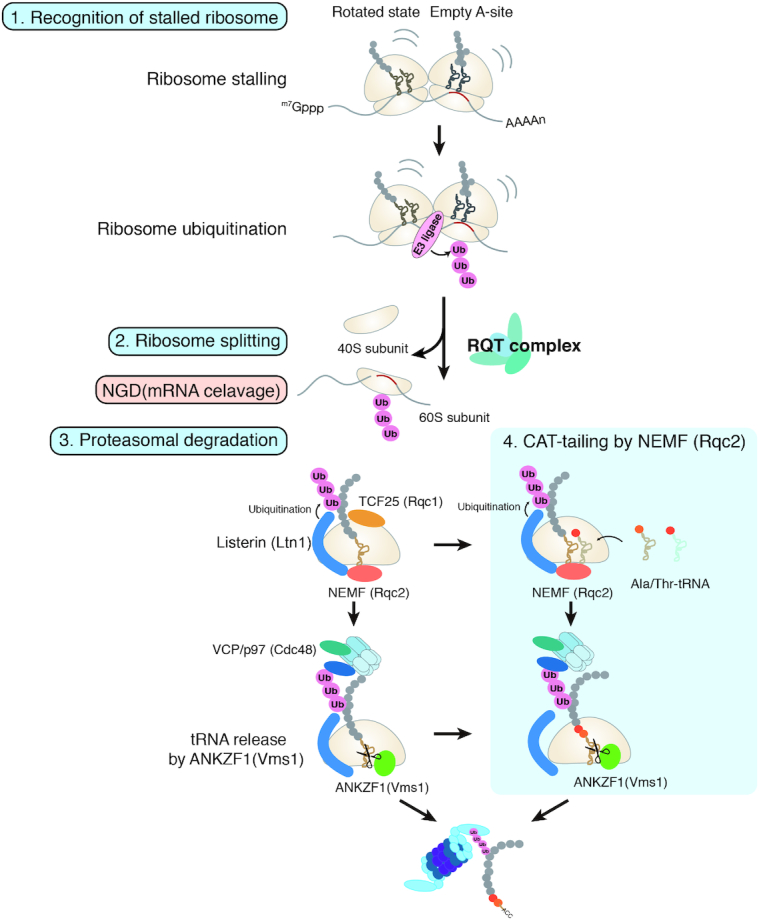
Quality control systems induced by ribosome stalling. RQC has three steps. Step 1: The abnormal stalled ribosome is recognized and ubiquitinated at one or more specific residues. Step 2: Ribosome ubiquitination induces subunit dissociation, which is a crucial event for Listerin (S. cerevisiae Ltn1)-dependent ubiquitination of the arrest products on the 60S large ribosomal subunit. The RQT complex is thought to recognize the ubiquitinated stalled ribosome and induce subunit dissociation in yeast. Step 3: Dissociation of the 40S subunits allows binding of 60S RNCs to NEMF (Rqc2), which recruits the E3 ubiquitin ligase Listerin (Ltn1). Subsequently, Listerin ubiquitinates the nascent polypeptide chains, targeting them for degradation. Rqc2 attaches CAT-tails (C-terminal alanyl/threonyl sequences) to stalled polypeptides. The ATPase VCP/p97 (Cdc48) forms a complex with the cofactors UFD1 (Ufd1) and NPLOC4 (Npl4) and unfolds ubiquitinated polypeptides, then extracts the peptidyl-tRNA from the 60S, thereby recruiting it to the 26S proteasome for degradation. Novel roles of ANKZF1 (Vms1)-mediated polypeptide release for proteasomal degradation: Rqc2 catalyzes the C-terminal extension of the stalled tRNA-bound peptides with CAT-tails through a non-canonical elongation reaction without mRNA. CAT-tails functionalize the carboxy termini of stalled polypeptides to drive their degradation on and off the ribosome. Vms1 interacts with the ribosomal 60S subunit to compete with Rqc2 and catalyze peptidyl-tRNA cleavage. Subsequently, tRNA nucleotidyl transferase 1 (TRNT1) is responsible for recycling of ANKZF1-cleaved tRNA fragments.
