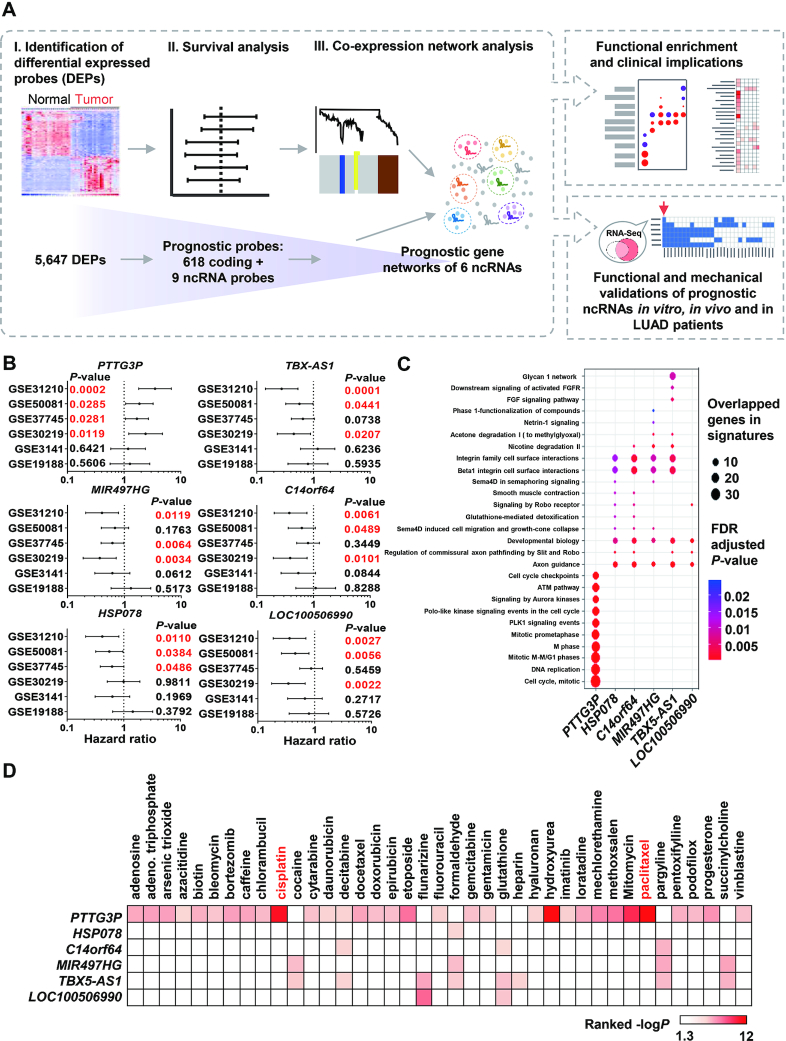
Figure 1.
Identification of prognostic ncRNAs in LUAD with predicted biological functions and drug responses. (A) A systematic pipeline was used to discover 5,647 differentially expressed probes (DEPs) and 627 prognostic probes, including nine noncoding probes, with log-rank tests based on multiple LUAD transcriptome datasets containing 10-year patient survival information. We identified six prognostic ncRNA-containing networks by WGCNA and then predicted their biological functions through WebGestalt. The validated and most promising ncRNAs were selected for functional and mechanistic studies of their roles in LUAD tumor progression and clinical intervention. (B) Survival analysis of ncRNAs in six LUAD transcriptomic datasets; a P-value lower than 0.05 is highlighted in red. (C) Pathway prediction in ncRNA modules (networks) by WebGestalt, and the enrichment results was generated using the clusterProfiler R package. The dot size indicates the number of overlapping genes, and the color indicates the significance of the adjusted P-value after scaling. (D) Drug association analysis of ncRNA-containing modules was performed in WebGestalt to determine their clinical effects. The color scale indicates the ranked –log P-value. The white box indicates that the P-value was over 0.05.