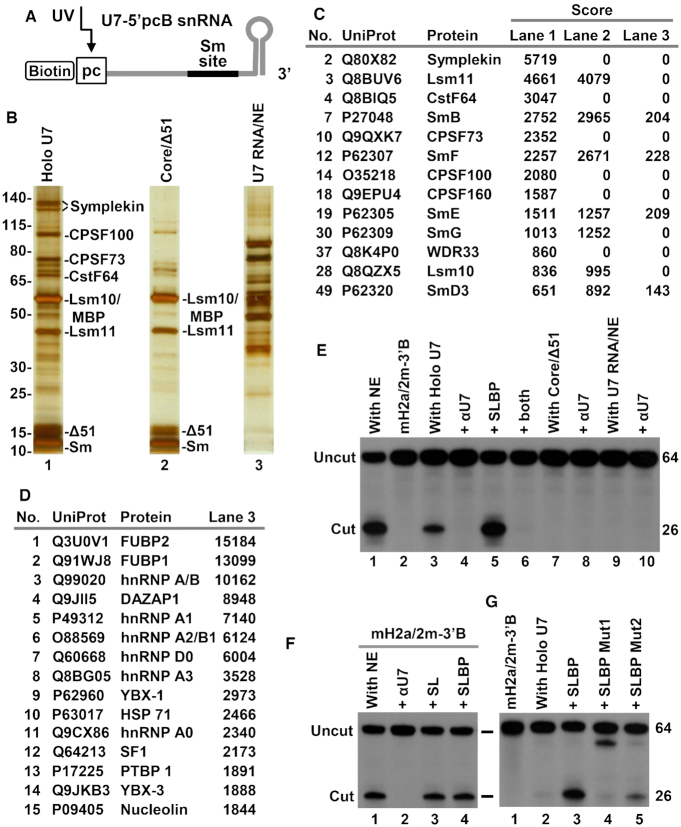
Figure 3.
Processing activity of purified semi-recombinant holo U7 snRNP. (A) Recombinant core U7 snRNP containing full length Lsm11 and SmB/Δ1–95 was assembled on synthetic U7–5′pcB snRNA tagged at the 5′ end with biotin and a photo-cleavable linker. (B) The assembled core was bound to Δ51 FLASH, immobilized on SA beads and incubated with mouse nuclear extract to recruit the HCC. The reconstituted semi-recombinant holo U7 snRNP was eluted to solution by UV irradiation and a fraction (10%) was analyzed by silver staining (lane 1). Silver staining of UV-eluted core U7 snRNP/Δ51 FLASH complex (no incubation with the nuclear extract) is shown in lane 2. Silver staining of UV-eluted U7–5′pcB snRNA (no Sm ring and FLASH) incubated with the same nuclear extract is shown in lane 3. (C) The same fraction (10%) of the three UV-eluted samples was analyzed by mass spectrometry. The table lists top scoring components (scores > 500) of semi-recombinant holo U7 snRNP (lane 1 in panel B). (D) Top scoring proteins identified in the UV-eluted sample of U7–5′pcB snRNA incubated with the nuclear extract (lane 3 in panel B). (E) A fraction (5%) of the UV-eluted samples shown in panel B were tested for the ability to support processing of mH2a/2m-3′B pre-mRNA either in the absence or in the presence of SLBP and/or an oligonucleotide blocking mouse U7 snRNA (αU7), as indicated at the top of each lane. (F) Processing of mH2a/2m-3′B pre-mRNA in a mouse nuclear extract alone (lane 1) or in the presence of SLBP, stem–loop RNA (SL) or αU7 oligonucleotide, as indicated at the top of each lane. (G) Processing of mH2a/2m-3′B pre-mRNA by a fraction (0.5%) of the UV-eluted semi-recombinant holo U7 snRNP alone (lane 2) or in the presence of WT SLBP (lane 3), or its mutant versions unable to support processing (lanes 4–5).